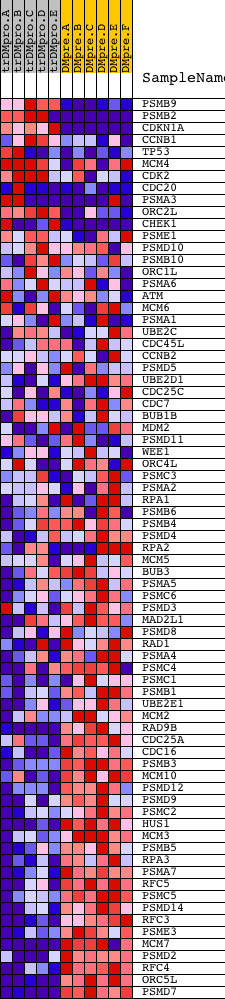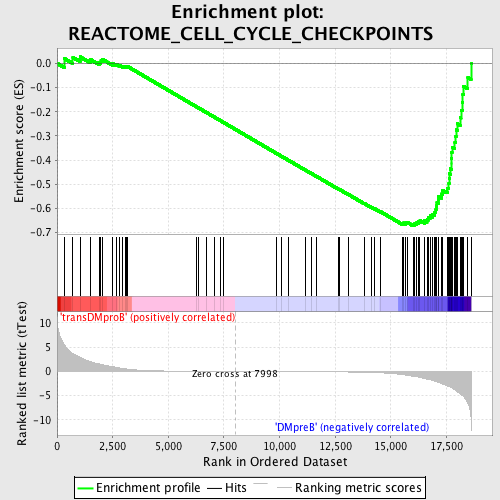
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | REACTOME_CELL_CYCLE_CHECKPOINTS |
| Enrichment Score (ES) | -0.6727944 |
| Normalized Enrichment Score (NES) | -1.6356586 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.021421678 |
| FWER p-Value | 0.403 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMB9 | 23021 | 323 | 5.459 | 0.0202 | No | ||
| 2 | PSMB2 | 2324 16078 | 711 | 3.670 | 0.0247 | No | ||
| 3 | CDKN1A | 4511 8729 | 1034 | 2.865 | 0.0271 | No | ||
| 4 | CCNB1 | 11201 21362 | 1487 | 2.009 | 0.0166 | No | ||
| 5 | TP53 | 20822 | 1912 | 1.507 | 0.0041 | No | ||
| 6 | MCM4 | 22655 1708 | 1971 | 1.440 | 0.0109 | No | ||
| 7 | CDK2 | 3438 3373 19592 3322 | 2032 | 1.374 | 0.0171 | No | ||
| 8 | CDC20 | 8421 8422 4227 | 2499 | 0.970 | -0.0013 | No | ||
| 9 | PSMA3 | 9632 5298 | 2673 | 0.808 | -0.0051 | No | ||
| 10 | ORC2L | 385 13949 | 2822 | 0.677 | -0.0084 | No | ||
| 11 | CHEK1 | 19181 3085 | 2960 | 0.576 | -0.0118 | No | ||
| 12 | PSME1 | 5300 9637 | 3070 | 0.516 | -0.0141 | No | ||
| 13 | PSMD10 | 2578 2597 24046 | 3127 | 0.485 | -0.0138 | No | ||
| 14 | PSMB10 | 5299 18761 | 3162 | 0.463 | -0.0124 | No | ||
| 15 | ORC1L | 327 16144 | 6252 | 0.027 | -0.1788 | No | ||
| 16 | PSMA6 | 21270 | 6335 | 0.025 | -0.1831 | No | ||
| 17 | ATM | 2976 19115 | 6719 | 0.018 | -0.2036 | No | ||
| 18 | MCM6 | 4000 13845 4119 | 7085 | 0.013 | -0.2232 | No | ||
| 19 | PSMA1 | 1627 17669 | 7334 | 0.009 | -0.2365 | No | ||
| 20 | UBE2C | 12714 | 7495 | 0.007 | -0.2451 | No | ||
| 21 | CDC45L | 22642 1752 | 9842 | -0.026 | -0.3714 | No | ||
| 22 | CCNB2 | 19067 | 10074 | -0.030 | -0.3837 | No | ||
| 23 | PSMD5 | 2774 14612 | 10407 | -0.035 | -0.4013 | No | ||
| 24 | UBE2D1 | 19738 | 11180 | -0.050 | -0.4426 | No | ||
| 25 | CDC25C | 23468 1954 1977 | 11439 | -0.056 | -0.4562 | No | ||
| 26 | CDC7 | 16765 3477 | 11672 | -0.062 | -0.4683 | No | ||
| 27 | BUB1B | 14908 | 12669 | -0.100 | -0.5213 | No | ||
| 28 | MDM2 | 19620 3327 | 12689 | -0.101 | -0.5216 | No | ||
| 29 | PSMD11 | 12772 7600 | 13078 | -0.123 | -0.5417 | No | ||
| 30 | WEE1 | 18127 | 13796 | -0.184 | -0.5791 | No | ||
| 31 | ORC4L | 11172 6460 | 14118 | -0.221 | -0.5949 | No | ||
| 32 | PSMC3 | 9636 | 14271 | -0.243 | -0.6014 | No | ||
| 33 | PSMA2 | 9631 | 14554 | -0.293 | -0.6146 | No | ||
| 34 | RPA1 | 20349 | 15517 | -0.662 | -0.6619 | No | ||
| 35 | PSMB6 | 9634 | 15579 | -0.698 | -0.6604 | No | ||
| 36 | PSMB4 | 15252 | 15677 | -0.765 | -0.6603 | No | ||
| 37 | PSMD4 | 15251 | 15753 | -0.824 | -0.6587 | No | ||
| 38 | RPA2 | 2330 16057 | 16016 | -1.005 | -0.6659 | Yes | ||
| 39 | MCM5 | 18564 | 16079 | -1.039 | -0.6620 | Yes | ||
| 40 | BUB3 | 18045 | 16170 | -1.110 | -0.6592 | Yes | ||
| 41 | PSMA5 | 6464 | 16241 | -1.174 | -0.6549 | Yes | ||
| 42 | PSMC6 | 7379 | 16310 | -1.237 | -0.6500 | Yes | ||
| 43 | PSMD3 | 5803 | 16523 | -1.485 | -0.6512 | Yes | ||
| 44 | MAD2L1 | 17422 | 16627 | -1.574 | -0.6459 | Yes | ||
| 45 | PSMD8 | 7166 | 16675 | -1.611 | -0.6374 | Yes | ||
| 46 | RAD1 | 22507 | 16765 | -1.702 | -0.6304 | Yes | ||
| 47 | PSMA4 | 11179 | 16871 | -1.842 | -0.6234 | Yes | ||
| 48 | PSMC4 | 2141 17915 | 16976 | -1.996 | -0.6152 | Yes | ||
| 49 | PSMC1 | 9635 | 17026 | -2.067 | -0.6036 | Yes | ||
| 50 | PSMB1 | 23118 | 17038 | -2.079 | -0.5898 | Yes | ||
| 51 | UBE2E1 | 10243 5817 | 17073 | -2.141 | -0.5769 | Yes | ||
| 52 | MCM2 | 17074 | 17133 | -2.214 | -0.5648 | Yes | ||
| 53 | RAD9B | 10541 | 17148 | -2.223 | -0.5502 | Yes | ||
| 54 | CDC25A | 8721 | 17285 | -2.488 | -0.5404 | Yes | ||
| 55 | CDC16 | 18676 | 17312 | -2.548 | -0.5242 | Yes | ||
| 56 | PSMB3 | 11180 | 17536 | -2.970 | -0.5158 | Yes | ||
| 57 | MCM10 | 14694 | 17609 | -3.085 | -0.4984 | Yes | ||
| 58 | PSMD12 | 20621 | 17621 | -3.111 | -0.4775 | Yes | ||
| 59 | PSMD9 | 3461 7393 | 17635 | -3.147 | -0.4565 | Yes | ||
| 60 | PSMC2 | 16909 | 17677 | -3.270 | -0.4362 | Yes | ||
| 61 | HUS1 | 9138 4890 | 17715 | -3.352 | -0.4150 | Yes | ||
| 62 | MCM3 | 13991 | 17716 | -3.357 | -0.3919 | Yes | ||
| 63 | PSMB5 | 9633 | 17739 | -3.411 | -0.3695 | Yes | ||
| 64 | RPA3 | 12667 | 17754 | -3.445 | -0.3465 | Yes | ||
| 65 | PSMA7 | 14318 | 17880 | -3.794 | -0.3271 | Yes | ||
| 66 | RFC5 | 13005 7791 | 17920 | -3.951 | -0.3019 | Yes | ||
| 67 | PSMC5 | 1348 20624 | 17937 | -3.998 | -0.2752 | Yes | ||
| 68 | PSMD14 | 15005 | 17987 | -4.160 | -0.2492 | Yes | ||
| 69 | RFC3 | 12786 | 18116 | -4.657 | -0.2239 | Yes | ||
| 70 | PSME3 | 20657 | 18155 | -4.800 | -0.1929 | Yes | ||
| 71 | MCM7 | 9372 3568 | 18211 | -4.973 | -0.1615 | Yes | ||
| 72 | PSMD2 | 10137 5724 | 18238 | -5.062 | -0.1280 | Yes | ||
| 73 | RFC4 | 1735 22627 | 18260 | -5.128 | -0.0938 | Yes | ||
| 74 | ORC5L | 11173 3595 | 18464 | -6.552 | -0.0595 | Yes | ||
| 75 | PSMD7 | 18752 3825 | 18607 | -9.809 | 0.0005 | Yes |