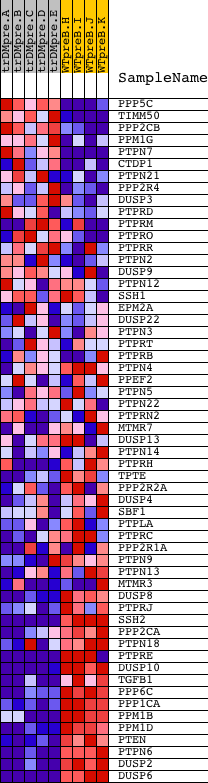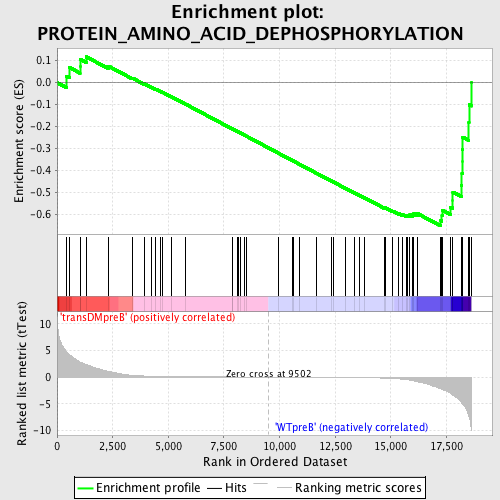
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | PROTEIN_AMINO_ACID_DEPHOSPHORYLATION |
| Enrichment Score (ES) | -0.6512217 |
| Normalized Enrichment Score (NES) | -1.4840369 |
| Nominal p-value | 0.006329114 |
| FDR q-value | 0.7043722 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PPP5C | 17952 | 429 | 4.765 | 0.0297 | No | ||
| 2 | TIMM50 | 17911 | 562 | 4.171 | 0.0688 | No | ||
| 3 | PPP2CB | 18636 | 1030 | 2.833 | 0.0751 | No | ||
| 4 | PPM1G | 16571 | 1034 | 2.823 | 0.1062 | No | ||
| 5 | PTPN7 | 11500 | 1312 | 2.372 | 0.1176 | No | ||
| 6 | CTDP1 | 23399 1969 | 2304 | 1.033 | 0.0756 | No | ||
| 7 | PTPN21 | 21009 | 3385 | 0.335 | 0.0212 | No | ||
| 8 | PPP2R4 | 15050 2910 | 3905 | 0.209 | -0.0045 | No | ||
| 9 | DUSP3 | 1358 13022 | 4241 | 0.159 | -0.0208 | No | ||
| 10 | PTPRD | 11554 5330 | 4431 | 0.138 | -0.0294 | No | ||
| 11 | PTPRM | 22904 | 4643 | 0.119 | -0.0395 | No | ||
| 12 | PTPRO | 1093 17256 | 4734 | 0.113 | -0.0431 | No | ||
| 13 | PTPRR | 3436 19876 | 5126 | 0.089 | -0.0631 | No | ||
| 14 | PTPN2 | 23414 | 5764 | 0.062 | -0.0968 | No | ||
| 15 | DUSP9 | 24307 | 7863 | 0.021 | -0.2096 | No | ||
| 16 | PTPN12 | 16597 3502 3586 3665 | 7872 | 0.021 | -0.2098 | No | ||
| 17 | SSH1 | 10540 | 8096 | 0.018 | -0.2216 | No | ||
| 18 | EPM2A | 20090 | 8151 | 0.018 | -0.2243 | No | ||
| 19 | DUSP22 | 466 21680 | 8256 | 0.016 | -0.2297 | No | ||
| 20 | PTPN3 | 9661 | 8441 | 0.014 | -0.2395 | No | ||
| 21 | PTPRT | 5333 | 8522 | 0.012 | -0.2437 | No | ||
| 22 | PTPRB | 19875 | 9953 | -0.006 | -0.3206 | No | ||
| 23 | PTPN4 | 13856 | 10562 | -0.014 | -0.3532 | No | ||
| 24 | PPEF2 | 16481 | 10613 | -0.014 | -0.3558 | No | ||
| 25 | PTPN5 | 17817 | 10905 | -0.018 | -0.3713 | No | ||
| 26 | PTPN22 | 1775 15470 | 11643 | -0.030 | -0.4106 | No | ||
| 27 | PTPRN2 | 9666 | 12319 | -0.043 | -0.4465 | No | ||
| 28 | MTMR7 | 12032 | 12403 | -0.045 | -0.4505 | No | ||
| 29 | DUSP13 | 21904 | 12980 | -0.061 | -0.4809 | No | ||
| 30 | PTPN14 | 14012 | 13388 | -0.076 | -0.5019 | No | ||
| 31 | PTPRH | 11861 | 13593 | -0.085 | -0.5120 | No | ||
| 32 | TPTE | 3875 3866 10615 3777 | 13835 | -0.100 | -0.5239 | No | ||
| 33 | PPP2R2A | 3222 21774 | 14724 | -0.187 | -0.5696 | No | ||
| 34 | DUSP4 | 18632 3820 | 14743 | -0.190 | -0.5685 | No | ||
| 35 | SBF1 | 13420 | 15096 | -0.257 | -0.5846 | No | ||
| 36 | PTPLA | 11403 6636 14680 | 15344 | -0.320 | -0.5944 | No | ||
| 37 | PTPRC | 5327 9662 | 15502 | -0.380 | -0.5986 | No | ||
| 38 | PPP2R1A | 11951 | 15700 | -0.464 | -0.6041 | No | ||
| 39 | PTPN9 | 19434 | 15760 | -0.492 | -0.6018 | No | ||
| 40 | PTPN13 | 16777 | 15857 | -0.548 | -0.6009 | No | ||
| 41 | MTMR3 | 7908 13193 | 15977 | -0.641 | -0.6002 | No | ||
| 42 | DUSP8 | 9493 | 16035 | -0.686 | -0.5957 | No | ||
| 43 | PTPRJ | 9664 | 16204 | -0.867 | -0.5951 | No | ||
| 44 | SSH2 | 6202 | 17246 | -2.211 | -0.6267 | Yes | ||
| 45 | PPP2CA | 20890 | 17299 | -2.304 | -0.6040 | Yes | ||
| 46 | PTPN18 | 14279 | 17310 | -2.320 | -0.5788 | Yes | ||
| 47 | PTPRE | 1662 18039 | 17672 | -2.996 | -0.5650 | Yes | ||
| 48 | DUSP10 | 4003 14016 | 17788 | -3.333 | -0.5343 | Yes | ||
| 49 | TGFB1 | 18332 | 17790 | -3.337 | -0.4973 | Yes | ||
| 50 | PPP6C | 7491 | 18185 | -4.785 | -0.4655 | Yes | ||
| 51 | PPP1CA | 9608 3756 914 909 | 18197 | -4.866 | -0.4122 | Yes | ||
| 52 | PPM1B | 5280 1567 5281 | 18219 | -4.963 | -0.3583 | Yes | ||
| 53 | PPM1D | 20721 | 18221 | -4.979 | -0.3031 | Yes | ||
| 54 | PTEN | 5305 | 18232 | -5.045 | -0.2478 | Yes | ||
| 55 | PTPN6 | 17002 | 18509 | -7.257 | -0.1822 | Yes | ||
| 56 | DUSP2 | 14865 | 18515 | -7.372 | -0.1007 | Yes | ||
| 57 | DUSP6 | 19891 3399 | 18611 | -9.573 | 0.0003 | Yes |