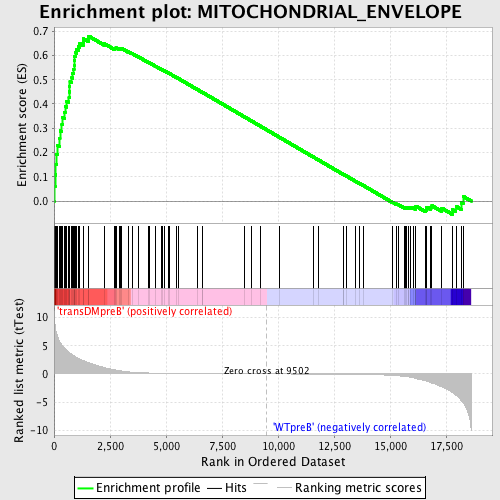
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | transDMpreB |
| GeneSet | MITOCHONDRIAL_ENVELOPE |
| Enrichment Score (ES) | 0.67969877 |
| Normalized Enrichment Score (NES) | 1.6278824 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.028046368 |
| FWER p-Value | 0.447 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TOMM22 | 5863 | 5 | 10.308 | 0.0602 | Yes | ||
| 2 | TIMM13 | 14974 | 46 | 8.588 | 0.1084 | Yes | ||
| 3 | TIMM10 | 11390 2158 | 80 | 7.684 | 0.1517 | Yes | ||
| 4 | ATP5G1 | 8636 | 101 | 7.285 | 0.1933 | Yes | ||
| 5 | TIMM17A | 13822 | 154 | 6.602 | 0.2292 | Yes | ||
| 6 | HTRA2 | 1122 17103 | 252 | 5.732 | 0.2576 | Yes | ||
| 7 | SLC25A11 | 20366 1368 | 263 | 5.654 | 0.2902 | Yes | ||
| 8 | TOMM34 | 14359 | 348 | 5.138 | 0.3158 | Yes | ||
| 9 | PHB2 | 17286 | 367 | 5.011 | 0.3443 | Yes | ||
| 10 | VDAC1 | 10282 | 461 | 4.601 | 0.3662 | Yes | ||
| 11 | BCS1L | 14223 | 494 | 4.470 | 0.3907 | Yes | ||
| 12 | TIMM50 | 17911 | 562 | 4.171 | 0.4116 | Yes | ||
| 13 | NDUFS3 | 7553 12683 | 663 | 3.847 | 0.4287 | Yes | ||
| 14 | ATP5G2 | 12610 | 681 | 3.781 | 0.4500 | Yes | ||
| 15 | UQCRB | 12545 | 697 | 3.715 | 0.4710 | Yes | ||
| 16 | CYCS | 8821 | 708 | 3.673 | 0.4920 | Yes | ||
| 17 | SLC25A22 | 12671 | 792 | 3.455 | 0.5077 | Yes | ||
| 18 | ATP5A1 | 23505 | 815 | 3.395 | 0.5265 | Yes | ||
| 19 | SLC25A15 | 356 3819 | 863 | 3.241 | 0.5429 | Yes | ||
| 20 | TIMM8B | 11391 | 890 | 3.180 | 0.5602 | Yes | ||
| 21 | BAX | 17832 | 898 | 3.158 | 0.5783 | Yes | ||
| 22 | ATP5G3 | 14558 | 918 | 3.114 | 0.5956 | Yes | ||
| 23 | ABCB6 | 13162 | 952 | 3.016 | 0.6115 | Yes | ||
| 24 | NDUFA9 | 12288 | 1004 | 2.896 | 0.6257 | Yes | ||
| 25 | NDUFS7 | 19946 | 1095 | 2.728 | 0.6369 | Yes | ||
| 26 | ATP5O | 22539 | 1145 | 2.647 | 0.6497 | Yes | ||
| 27 | SLC25A3 | 19644 | 1296 | 2.390 | 0.6557 | Yes | ||
| 28 | ACN9 | 12920 | 1318 | 2.365 | 0.6684 | Yes | ||
| 29 | NDUFV1 | 23953 | 1528 | 2.008 | 0.6689 | Yes | ||
| 30 | SDHD | 19125 | 1545 | 1.990 | 0.6797 | Yes | ||
| 31 | SLC25A1 | 22645 | 2243 | 1.101 | 0.6485 | No | ||
| 32 | RHOT1 | 12227 | 2713 | 0.733 | 0.6275 | No | ||
| 33 | NDUFAB1 | 7667 | 2732 | 0.718 | 0.6308 | No | ||
| 34 | SURF1 | 14645 | 2799 | 0.660 | 0.6311 | No | ||
| 35 | ATP5D | 19949 | 2903 | 0.583 | 0.6289 | No | ||
| 36 | TIMM9 | 19686 | 2960 | 0.553 | 0.6292 | No | ||
| 37 | VDAC3 | 10284 | 3011 | 0.512 | 0.6295 | No | ||
| 38 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 3317 | 0.365 | 0.6152 | No | ||
| 39 | VDAC2 | 22081 | 3482 | 0.305 | 0.6081 | No | ||
| 40 | ATP5E | 14321 | 3762 | 0.235 | 0.5944 | No | ||
| 41 | MPV17 | 9407 3505 3543 | 4216 | 0.162 | 0.5709 | No | ||
| 42 | MTX2 | 14980 | 4235 | 0.160 | 0.5709 | No | ||
| 43 | ATP5J | 880 22555 | 4543 | 0.127 | 0.5551 | No | ||
| 44 | NDUFA2 | 9450 | 4786 | 0.110 | 0.5426 | No | ||
| 45 | HSD3B2 | 4873 4874 15229 4875 9126 | 4849 | 0.106 | 0.5399 | No | ||
| 46 | CENTA2 | 20748 | 4949 | 0.099 | 0.5352 | No | ||
| 47 | MFN2 | 15678 2417 | 5096 | 0.091 | 0.5278 | No | ||
| 48 | TIMM23 | 7004 5768 | 5150 | 0.088 | 0.5255 | No | ||
| 49 | COX15 | 5931 | 5462 | 0.074 | 0.5091 | No | ||
| 50 | BNIP3 | 17581 | 5548 | 0.070 | 0.5049 | No | ||
| 51 | NDUFS1 | 13940 | 6414 | 0.046 | 0.4585 | No | ||
| 52 | HCCS | 9079 4842 | 6628 | 0.042 | 0.4473 | No | ||
| 53 | UCP3 | 3972 18175 | 6633 | 0.042 | 0.4473 | No | ||
| 54 | UQCRC1 | 10259 | 8509 | 0.013 | 0.3462 | No | ||
| 55 | IMMT | 367 8029 | 8809 | 0.009 | 0.3301 | No | ||
| 56 | SDHA | 12436 | 9232 | 0.004 | 0.3073 | No | ||
| 57 | OPA1 | 13169 22800 7894 | 10064 | -0.007 | 0.2625 | No | ||
| 58 | PPOX | 13761 4104 | 11559 | -0.028 | 0.1821 | No | ||
| 59 | PHB | 9558 | 11782 | -0.032 | 0.1703 | No | ||
| 60 | HSD3B1 | 15227 | 12908 | -0.058 | 0.1099 | No | ||
| 61 | MAOB | 24182 2556 | 12918 | -0.058 | 0.1098 | No | ||
| 62 | NDUFA1 | 24170 | 13060 | -0.063 | 0.1025 | No | ||
| 63 | GATM | 14451 | 13453 | -0.078 | 0.0818 | No | ||
| 64 | ATP5B | 19846 | 13631 | -0.088 | 0.0728 | No | ||
| 65 | COX6B2 | 17983 4110 | 13818 | -0.099 | 0.0633 | No | ||
| 66 | NDUFS2 | 4089 13760 | 15119 | -0.261 | -0.0053 | No | ||
| 67 | RHOT2 | 1533 10073 | 15275 | -0.302 | -0.0119 | No | ||
| 68 | NNT | 3273 5181 9471 3238 | 15351 | -0.322 | -0.0141 | No | ||
| 69 | MRPL32 | 7961 | 15632 | -0.435 | -0.0266 | No | ||
| 70 | AIFM2 | 3320 3407 20006 3317 | 15689 | -0.458 | -0.0270 | No | ||
| 71 | NDUFA13 | 7400 | 15718 | -0.474 | -0.0257 | No | ||
| 72 | NDUFS4 | 9452 5157 3173 | 15820 | -0.525 | -0.0281 | No | ||
| 73 | RAB11FIP5 | 17089 | 15833 | -0.534 | -0.0256 | No | ||
| 74 | ALAS2 | 24229 | 15930 | -0.601 | -0.0272 | No | ||
| 75 | CASP7 | 23826 | 16022 | -0.678 | -0.0282 | No | ||
| 76 | ATP5C1 | 8635 | 16131 | -0.780 | -0.0294 | No | ||
| 77 | PMPCA | 12421 7350 | 16138 | -0.790 | -0.0251 | No | ||
| 78 | OGDH | 9502 1412 | 16147 | -0.807 | -0.0208 | No | ||
| 79 | UQCRH | 12378 | 16588 | -1.209 | -0.0375 | No | ||
| 80 | RAF1 | 17035 | 16625 | -1.259 | -0.0320 | No | ||
| 81 | NDUFS8 | 23950 | 16626 | -1.261 | -0.0246 | No | ||
| 82 | ABCF2 | 11338 6580 6579 | 16794 | -1.501 | -0.0248 | No | ||
| 83 | NDUFA6 | 12473 | 16866 | -1.614 | -0.0192 | No | ||
| 84 | HADHB | 3514 10527 | 17305 | -2.309 | -0.0293 | No | ||
| 85 | ABCB7 | 24085 | 17789 | -3.335 | -0.0358 | No | ||
| 86 | TIMM17B | 24390 | 17940 | -3.775 | -0.0218 | No | ||
| 87 | FIS1 | 16669 3579 | 18178 | -4.737 | -0.0068 | No | ||
| 88 | MCL1 | 15502 | 18260 | -5.175 | 0.0192 | No |

