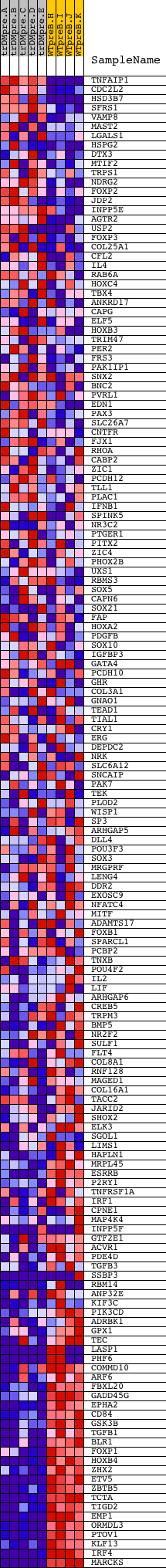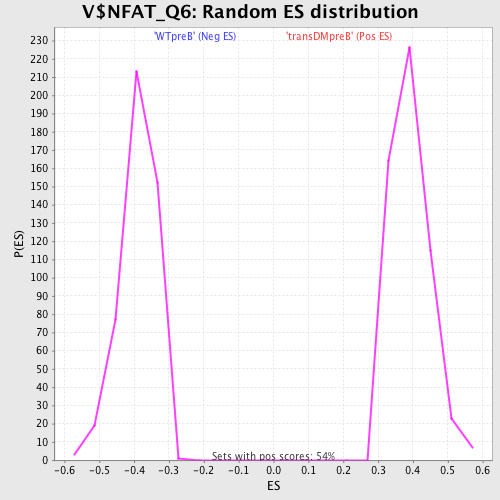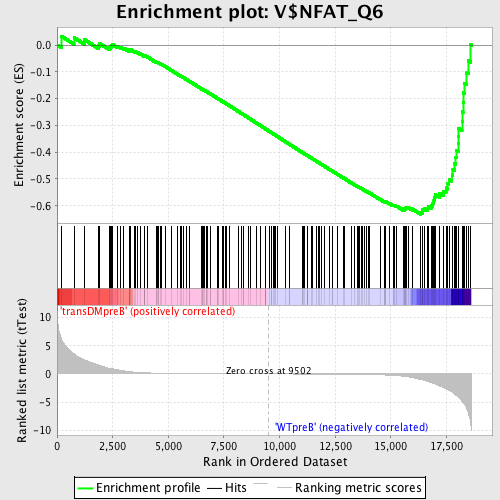
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | V$NFAT_Q6 |
| Enrichment Score (ES) | -0.6326506 |
| Normalized Enrichment Score (NES) | -1.6291189 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.054269306 |
| FWER p-Value | 0.187 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TNFAIP1 | 20333 | 196 | 6.200 | 0.0335 | No | ||
| 2 | CDC2L2 | 15964 2419 | 760 | 3.548 | 0.0283 | No | ||
| 3 | HSD3B7 | 4158 | 1217 | 2.505 | 0.0214 | No | ||
| 4 | SFRS1 | 8492 | 1857 | 1.596 | -0.0019 | No | ||
| 5 | VAMP8 | 1034 17117 | 1901 | 1.536 | 0.0067 | No | ||
| 6 | MAST2 | 9423 | 2345 | 1.005 | -0.0101 | No | ||
| 7 | LGALS1 | 22429 | 2410 | 0.972 | -0.0067 | No | ||
| 8 | HSPG2 | 4884 16023 | 2429 | 0.957 | -0.0008 | No | ||
| 9 | DTX3 | 19607 | 2502 | 0.895 | 0.0016 | No | ||
| 10 | MTIF2 | 13354 | 2726 | 0.722 | -0.0053 | No | ||
| 11 | TRPS1 | 8195 | 2846 | 0.627 | -0.0073 | No | ||
| 12 | NDRG2 | 21843 | 2986 | 0.535 | -0.0110 | No | ||
| 13 | FOXP2 | 4312 17523 8523 | 3234 | 0.394 | -0.0216 | No | ||
| 14 | JDP2 | 21201 | 3246 | 0.390 | -0.0194 | No | ||
| 15 | INPP5E | 14650 | 3249 | 0.390 | -0.0168 | No | ||
| 16 | AGTR2 | 24365 | 3309 | 0.368 | -0.0173 | No | ||
| 17 | USP2 | 19480 3052 3043 | 3489 | 0.303 | -0.0249 | No | ||
| 18 | FOXP3 | 9804 5426 | 3523 | 0.292 | -0.0246 | No | ||
| 19 | COL25A1 | 8056 1837 1844 1915 13376 | 3618 | 0.268 | -0.0278 | No | ||
| 20 | CFL2 | 2155 21069 | 3742 | 0.239 | -0.0327 | No | ||
| 21 | IL4 | 9174 | 3912 | 0.208 | -0.0404 | No | ||
| 22 | RAB6A | 18173 3905 | 3919 | 0.207 | -0.0393 | No | ||
| 23 | HOXC4 | 4864 | 3935 | 0.203 | -0.0386 | No | ||
| 24 | TBX4 | 5633 | 4061 | 0.182 | -0.0441 | No | ||
| 25 | ANKRD17 | 13496 3474 8170 | 4466 | 0.135 | -0.0650 | No | ||
| 26 | CAPG | 17411 | 4488 | 0.133 | -0.0652 | No | ||
| 27 | ELF5 | 14936 2913 | 4507 | 0.131 | -0.0653 | No | ||
| 28 | HOXB3 | 20685 | 4551 | 0.127 | -0.0667 | No | ||
| 29 | TRIM47 | 10130 | 4568 | 0.126 | -0.0667 | No | ||
| 30 | PER2 | 3940 13881 | 4638 | 0.120 | -0.0695 | No | ||
| 31 | FRS3 | 23209 | 4713 | 0.115 | -0.0727 | No | ||
| 32 | PAK1IP1 | 21660 | 4874 | 0.105 | -0.0807 | No | ||
| 33 | SNX2 | 1974 23554 12585 | 5138 | 0.089 | -0.0943 | No | ||
| 34 | BNC2 | 15850 | 5408 | 0.076 | -0.1083 | No | ||
| 35 | PVRL1 | 3006 19482 12211 7192 | 5564 | 0.069 | -0.1162 | No | ||
| 36 | EDN1 | 21658 | 5572 | 0.069 | -0.1161 | No | ||
| 37 | PAX3 | 13910 | 5683 | 0.065 | -0.1216 | No | ||
| 38 | SLC26A7 | 5538 2372 | 5833 | 0.060 | -0.1292 | No | ||
| 39 | CNTFR | 2515 15906 | 5947 | 0.057 | -0.1350 | No | ||
| 40 | FJX1 | 4725 | 5964 | 0.057 | -0.1354 | No | ||
| 41 | RHOA | 8624 4409 4410 | 6494 | 0.044 | -0.1638 | No | ||
| 42 | CABP2 | 23765 | 6516 | 0.044 | -0.1646 | No | ||
| 43 | ZIC1 | 19040 | 6531 | 0.044 | -0.1650 | No | ||
| 44 | PCDH12 | 7005 | 6581 | 0.043 | -0.1674 | No | ||
| 45 | TLL1 | 10188 | 6641 | 0.041 | -0.1703 | No | ||
| 46 | PLAC1 | 12079 | 6697 | 0.040 | -0.1730 | No | ||
| 47 | IFNB1 | 15846 | 6703 | 0.040 | -0.1730 | No | ||
| 48 | SPINK5 | 23569 | 6759 | 0.039 | -0.1757 | No | ||
| 49 | NR3C2 | 18562 13798 | 6899 | 0.037 | -0.1829 | No | ||
| 50 | PTGER1 | 5309 | 6901 | 0.037 | -0.1827 | No | ||
| 51 | PITX2 | 15424 1878 | 7188 | 0.031 | -0.1980 | No | ||
| 52 | ZIC4 | 5992 | 7197 | 0.031 | -0.1982 | No | ||
| 53 | PHOX2B | 16524 | 7270 | 0.030 | -0.2019 | No | ||
| 54 | UXS1 | 13965 | 7434 | 0.027 | -0.2105 | No | ||
| 55 | RBMS3 | 18973 | 7457 | 0.027 | -0.2115 | No | ||
| 56 | SOX5 | 9850 1044 5480 16937 | 7475 | 0.027 | -0.2123 | No | ||
| 57 | CAPN6 | 24041 | 7554 | 0.026 | -0.2163 | No | ||
| 58 | SOX21 | 27 10278 2778 | 7618 | 0.025 | -0.2196 | No | ||
| 59 | FAP | 14577 | 7752 | 0.023 | -0.2266 | No | ||
| 60 | HOXA2 | 17151 | 8159 | 0.017 | -0.2485 | No | ||
| 61 | PDGFB | 22199 2311 | 8287 | 0.016 | -0.2552 | No | ||
| 62 | SOX10 | 22211 | 8391 | 0.014 | -0.2607 | No | ||
| 63 | IGFBP3 | 4898 | 8398 | 0.014 | -0.2609 | No | ||
| 64 | GATA4 | 21792 4755 | 8608 | 0.011 | -0.2722 | No | ||
| 65 | PCDH10 | 5227 9534 1893 | 8692 | 0.010 | -0.2766 | No | ||
| 66 | GHR | 22336 | 8939 | 0.007 | -0.2899 | No | ||
| 67 | COL3A1 | 14253 8765 | 9158 | 0.004 | -0.3017 | No | ||
| 68 | GNAO1 | 4785 3829 | 9360 | 0.002 | -0.3125 | No | ||
| 69 | TEAD1 | 18121 | 9364 | 0.002 | -0.3127 | No | ||
| 70 | TIAL1 | 996 5753 | 9368 | 0.002 | -0.3128 | No | ||
| 71 | CRY1 | 19662 | 9526 | -0.000 | -0.3214 | No | ||
| 72 | ERG | 1686 8915 | 9637 | -0.002 | -0.3273 | No | ||
| 73 | DEPDC2 | 14295 | 9655 | -0.002 | -0.3282 | No | ||
| 74 | NRK | 6559 | 9714 | -0.003 | -0.3313 | No | ||
| 75 | SLC6A12 | 17299 | 9751 | -0.003 | -0.3333 | No | ||
| 76 | SNCAIP | 12589 | 9831 | -0.005 | -0.3375 | No | ||
| 77 | PAK7 | 14417 | 9893 | -0.005 | -0.3408 | No | ||
| 78 | TEK | 16175 | 10277 | -0.010 | -0.3614 | No | ||
| 79 | PLOD2 | 19352 | 10435 | -0.012 | -0.3699 | No | ||
| 80 | WISP1 | 22463 2289 | 10444 | -0.012 | -0.3702 | No | ||
| 81 | SP3 | 5483 | 11020 | -0.020 | -0.4012 | No | ||
| 82 | ARHGAP5 | 4412 8625 | 11027 | -0.020 | -0.4014 | No | ||
| 83 | DLL4 | 7040 | 11060 | -0.021 | -0.4030 | No | ||
| 84 | POU3F3 | 14261 | 11115 | -0.021 | -0.4058 | No | ||
| 85 | SOX3 | 24145 | 11232 | -0.023 | -0.4119 | No | ||
| 86 | MRGPRF | 17995 | 11251 | -0.023 | -0.4127 | No | ||
| 87 | LENG4 | 12886 23773 | 11257 | -0.023 | -0.4128 | No | ||
| 88 | DDR2 | 13767 | 11439 | -0.026 | -0.4224 | No | ||
| 89 | EXOSC9 | 15610 | 11478 | -0.027 | -0.4243 | No | ||
| 90 | NFATC4 | 22002 | 11666 | -0.030 | -0.4342 | No | ||
| 91 | MITF | 17349 | 11768 | -0.032 | -0.4395 | No | ||
| 92 | ADAMTS17 | 598 | 11793 | -0.032 | -0.4405 | No | ||
| 93 | FOXB1 | 12254 19069 | 11903 | -0.034 | -0.4462 | No | ||
| 94 | SPARCL1 | 16457 | 12009 | -0.036 | -0.4516 | No | ||
| 95 | PCBP2 | 9533 | 12237 | -0.041 | -0.4636 | No | ||
| 96 | TNXB | 1557 8174 878 8175 | 12368 | -0.044 | -0.4704 | No | ||
| 97 | POU4F2 | 9603 5276 | 12396 | -0.045 | -0.4715 | No | ||
| 98 | IL2 | 15354 | 12608 | -0.050 | -0.4826 | No | ||
| 99 | LIF | 956 667 20960 | 12854 | -0.056 | -0.4955 | No | ||
| 100 | ARHGAP6 | 24207 | 12930 | -0.059 | -0.4991 | No | ||
| 101 | CREB5 | 10551 | 13238 | -0.070 | -0.5152 | No | ||
| 102 | TRPM3 | 23904 10357 5927 | 13369 | -0.075 | -0.5217 | No | ||
| 103 | BMP5 | 19376 | 13482 | -0.079 | -0.5272 | No | ||
| 104 | NR2F2 | 8619 409 | 13557 | -0.083 | -0.5307 | No | ||
| 105 | SULF1 | 6285 | 13602 | -0.086 | -0.5324 | No | ||
| 106 | FLT4 | 20904 1461 | 13679 | -0.090 | -0.5359 | No | ||
| 107 | COL8A1 | 4548 | 13728 | -0.094 | -0.5379 | No | ||
| 108 | RNF128 | 24236 2641 | 13825 | -0.099 | -0.5424 | No | ||
| 109 | MAGED1 | 24108 | 13892 | -0.104 | -0.5452 | No | ||
| 110 | COL16A1 | 16066 | 13977 | -0.110 | -0.5490 | No | ||
| 111 | TACC2 | 18052 2534 2234 2435 2350 | 14051 | -0.115 | -0.5521 | No | ||
| 112 | JARID2 | 9200 | 14516 | -0.158 | -0.5761 | No | ||
| 113 | SHOX2 | 1802 5432 | 14711 | -0.185 | -0.5853 | No | ||
| 114 | ELK3 | 4663 94 3388 | 14736 | -0.189 | -0.5853 | No | ||
| 115 | SGOL1 | 7810 7809 13030 | 14763 | -0.192 | -0.5853 | No | ||
| 116 | LIMS1 | 8493 4290 | 14778 | -0.195 | -0.5847 | No | ||
| 117 | HAPLN1 | 21592 | 14944 | -0.225 | -0.5920 | No | ||
| 118 | MRPL45 | 20680 | 15101 | -0.258 | -0.5986 | No | ||
| 119 | ESRRB | 21196 | 15122 | -0.262 | -0.5978 | No | ||
| 120 | P2RY1 | 15581 | 15154 | -0.268 | -0.5976 | No | ||
| 121 | TNFRSF1A | 1181 10206 | 15257 | -0.296 | -0.6010 | No | ||
| 122 | IRF1 | 1336 1258 1433 9182 | 15585 | -0.416 | -0.6158 | No | ||
| 123 | CPNE1 | 11186 | 15613 | -0.426 | -0.6142 | No | ||
| 124 | MAP4K4 | 11241 | 15629 | -0.433 | -0.6120 | No | ||
| 125 | INPP5F | 18055 3877 | 15664 | -0.448 | -0.6106 | No | ||
| 126 | GTF2E1 | 22597 | 15679 | -0.454 | -0.6081 | No | ||
| 127 | ACVR1 | 4334 | 15722 | -0.475 | -0.6070 | No | ||
| 128 | PDE4D | 10722 6235 | 15809 | -0.523 | -0.6080 | No | ||
| 129 | TGFB3 | 10161 | 15964 | -0.628 | -0.6118 | No | ||
| 130 | SSBP3 | 2439 7814 2431 | 16349 | -0.999 | -0.6255 | Yes | ||
| 131 | RBM14 | 3673 12087 | 16416 | -1.043 | -0.6217 | Yes | ||
| 132 | ANP32E | 15500 | 16431 | -1.052 | -0.6150 | Yes | ||
| 133 | KIF3C | 9220 4954 | 16514 | -1.135 | -0.6113 | Yes | ||
| 134 | PIK3CD | 9563 | 16667 | -1.326 | -0.6101 | Yes | ||
| 135 | ADRBK1 | 23960 | 16689 | -1.352 | -0.6016 | Yes | ||
| 136 | GPX1 | 19310 | 16820 | -1.535 | -0.5978 | Yes | ||
| 137 | TEC | 16514 | 16891 | -1.651 | -0.5898 | Yes | ||
| 138 | LASP1 | 4986 4985 1248 | 16935 | -1.719 | -0.5799 | Yes | ||
| 139 | PHF6 | 2648 24340 | 16954 | -1.730 | -0.5685 | Yes | ||
| 140 | COMMD10 | 23561 | 16987 | -1.770 | -0.5577 | Yes | ||
| 141 | ARF6 | 21252 | 17192 | -2.126 | -0.5536 | Yes | ||
| 142 | FBXL20 | 13010 | 17370 | -2.421 | -0.5460 | Yes | ||
| 143 | GADD45G | 21637 | 17483 | -2.597 | -0.5335 | Yes | ||
| 144 | EPHA2 | 16006 | 17560 | -2.758 | -0.5180 | Yes | ||
| 145 | CD84 | 14049 | 17614 | -2.864 | -0.5005 | Yes | ||
| 146 | GSK3B | 22761 | 17749 | -3.186 | -0.4851 | Yes | ||
| 147 | TGFB1 | 18332 | 17790 | -3.337 | -0.4635 | Yes | ||
| 148 | BLR1 | 19145 3154 | 17862 | -3.555 | -0.4421 | Yes | ||
| 149 | FOXP1 | 4242 | 17922 | -3.738 | -0.4186 | Yes | ||
| 150 | HOXB4 | 20686 | 17960 | -3.831 | -0.3934 | Yes | ||
| 151 | ZHX2 | 11842 | 18038 | -4.076 | -0.3685 | Yes | ||
| 152 | ETV5 | 22630 | 18044 | -4.102 | -0.3396 | Yes | ||
| 153 | ZBTB5 | 15891 | 18056 | -4.139 | -0.3107 | Yes | ||
| 154 | TCTA | 4167 | 18202 | -4.896 | -0.2838 | Yes | ||
| 155 | TIGD2 | 17429 | 18209 | -4.913 | -0.2491 | Yes | ||
| 156 | EMP1 | 17260 | 18249 | -5.131 | -0.2147 | Yes | ||
| 157 | ORMDL3 | 12386 | 18267 | -5.225 | -0.1784 | Yes | ||
| 158 | PTOV1 | 13522 | 18312 | -5.412 | -0.1423 | Yes | ||
| 159 | KLF13 | 17807 | 18416 | -6.159 | -0.1040 | Yes | ||
| 160 | IRF4 | 21679 | 18488 | -6.955 | -0.0584 | Yes | ||
| 161 | MARCKS | 9331 | 18599 | -9.166 | 0.0009 | Yes |