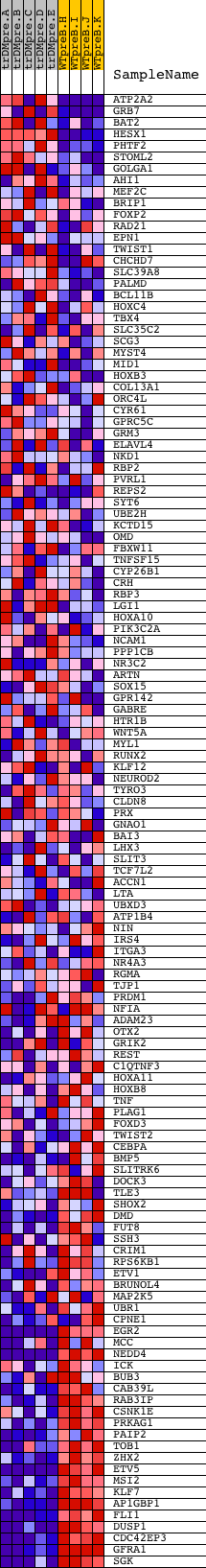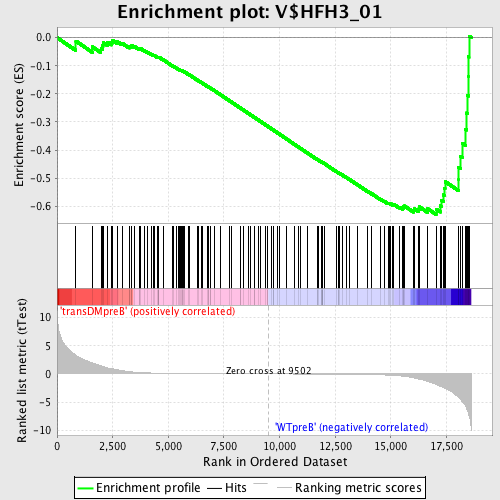
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | V$HFH3_01 |
| Enrichment Score (ES) | -0.6295045 |
| Normalized Enrichment Score (NES) | -1.5932925 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.060152844 |
| FWER p-Value | 0.307 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP2A2 | 4421 3481 | 846 | 3.299 | -0.0124 | No | ||
| 2 | GRB7 | 20673 | 1581 | 1.934 | -0.0325 | No | ||
| 3 | BAT2 | 23006 | 1972 | 1.436 | -0.0390 | No | ||
| 4 | HESX1 | 22070 | 2036 | 1.352 | -0.0287 | No | ||
| 5 | PHTF2 | 16599 | 2063 | 1.321 | -0.0168 | No | ||
| 6 | STOML2 | 15902 | 2264 | 1.073 | -0.0167 | No | ||
| 7 | GOLGA1 | 14595 | 2439 | 0.947 | -0.0165 | No | ||
| 8 | AHI1 | 20077 | 2488 | 0.906 | -0.0100 | No | ||
| 9 | MEF2C | 3204 9378 | 2707 | 0.739 | -0.0143 | No | ||
| 10 | BRIP1 | 20311 | 2922 | 0.572 | -0.0201 | No | ||
| 11 | FOXP2 | 4312 17523 8523 | 3234 | 0.394 | -0.0329 | No | ||
| 12 | RAD21 | 22298 | 3266 | 0.383 | -0.0307 | No | ||
| 13 | EPN1 | 18401 | 3336 | 0.359 | -0.0308 | No | ||
| 14 | TWIST1 | 21291 | 3357 | 0.347 | -0.0283 | No | ||
| 15 | CHCHD7 | 16280 | 3470 | 0.308 | -0.0313 | No | ||
| 16 | SLC39A8 | 12547 1776 | 3697 | 0.249 | -0.0410 | No | ||
| 17 | PALMD | 15181 | 3713 | 0.246 | -0.0393 | No | ||
| 18 | BCL11B | 7190 | 3740 | 0.240 | -0.0383 | No | ||
| 19 | HOXC4 | 4864 | 3935 | 0.203 | -0.0467 | No | ||
| 20 | TBX4 | 5633 | 4061 | 0.182 | -0.0516 | No | ||
| 21 | SLC35C2 | 10463 14339 | 4238 | 0.160 | -0.0595 | No | ||
| 22 | SCG3 | 19061 | 4326 | 0.149 | -0.0627 | No | ||
| 23 | MYST4 | 7027 7026 12014 2739 | 4364 | 0.145 | -0.0633 | No | ||
| 24 | MID1 | 5097 5098 | 4512 | 0.131 | -0.0699 | No | ||
| 25 | HOXB3 | 20685 | 4551 | 0.127 | -0.0707 | No | ||
| 26 | COL13A1 | 8763 | 4555 | 0.127 | -0.0695 | No | ||
| 27 | ORC4L | 11172 6460 | 4571 | 0.126 | -0.0691 | No | ||
| 28 | CYR61 | 9159 15145 9311 5024 | 4769 | 0.111 | -0.0786 | No | ||
| 29 | GPRC5C | 12864 | 5208 | 0.085 | -0.1014 | No | ||
| 30 | GRM3 | 16929 | 5231 | 0.084 | -0.1018 | No | ||
| 31 | ELAVL4 | 15805 4889 9137 | 5370 | 0.077 | -0.1084 | No | ||
| 32 | NKD1 | 8228 | 5466 | 0.074 | -0.1128 | No | ||
| 33 | RBP2 | 19347 | 5521 | 0.071 | -0.1150 | No | ||
| 34 | PVRL1 | 3006 19482 12211 7192 | 5564 | 0.069 | -0.1166 | No | ||
| 35 | REPS2 | 24018 | 5571 | 0.069 | -0.1162 | No | ||
| 36 | SYT6 | 13437 7042 12040 | 5616 | 0.067 | -0.1179 | No | ||
| 37 | UBE2H | 5823 5822 | 5656 | 0.066 | -0.1194 | No | ||
| 38 | KCTD15 | 17864 | 5681 | 0.065 | -0.1200 | No | ||
| 39 | OMD | 21643 | 5730 | 0.064 | -0.1220 | No | ||
| 40 | FBXW11 | 20926 | 5922 | 0.058 | -0.1317 | No | ||
| 41 | TNFSF15 | 15863 | 5968 | 0.057 | -0.1336 | No | ||
| 42 | CYP26B1 | 17093 | 6292 | 0.048 | -0.1505 | No | ||
| 43 | CRH | 8784 | 6337 | 0.047 | -0.1524 | No | ||
| 44 | RBP3 | 22047 | 6484 | 0.045 | -0.1599 | No | ||
| 45 | LGI1 | 23867 | 6519 | 0.044 | -0.1613 | No | ||
| 46 | HOXA10 | 17145 989 | 6757 | 0.039 | -0.1737 | No | ||
| 47 | PIK3C2A | 17667 | 6785 | 0.038 | -0.1748 | No | ||
| 48 | NCAM1 | 5149 | 6809 | 0.038 | -0.1756 | No | ||
| 49 | PPP1CB | 5282 3529 3504 | 6889 | 0.037 | -0.1795 | No | ||
| 50 | NR3C2 | 18562 13798 | 6899 | 0.037 | -0.1797 | No | ||
| 51 | ARTN | 15783 2430 2532 | 7056 | 0.034 | -0.1877 | No | ||
| 52 | SOX15 | 9848 | 7332 | 0.029 | -0.2023 | No | ||
| 53 | GPR142 | 20604 | 7736 | 0.023 | -0.2239 | No | ||
| 54 | GABRE | 24140 | 7825 | 0.022 | -0.2284 | No | ||
| 55 | HTR1B | 19051 | 8221 | 0.017 | -0.2496 | No | ||
| 56 | WNT5A | 22066 5880 | 8233 | 0.016 | -0.2501 | No | ||
| 57 | MYL1 | 4015 | 8369 | 0.015 | -0.2572 | No | ||
| 58 | RUNX2 | 4480 8700 | 8624 | 0.011 | -0.2708 | No | ||
| 59 | KLF12 | 4960 2637 21732 9228 | 8710 | 0.010 | -0.2753 | No | ||
| 60 | NEUROD2 | 20262 | 8856 | 0.008 | -0.2831 | No | ||
| 61 | TYRO3 | 5811 | 8858 | 0.008 | -0.2831 | No | ||
| 62 | CLDN8 | 22549 1747 | 9068 | 0.006 | -0.2943 | No | ||
| 63 | PRX | 3729 9628 | 9131 | 0.005 | -0.2976 | No | ||
| 64 | GNAO1 | 4785 3829 | 9360 | 0.002 | -0.3099 | No | ||
| 65 | BAI3 | 5580 | 9437 | 0.001 | -0.3140 | No | ||
| 66 | LHX3 | 2761 4996 | 9466 | 0.000 | -0.3155 | No | ||
| 67 | SLIT3 | 20925 | 9626 | -0.002 | -0.3241 | No | ||
| 68 | TCF7L2 | 10048 5646 | 9716 | -0.003 | -0.3289 | No | ||
| 69 | ACCN1 | 20327 1194 | 9718 | -0.003 | -0.3289 | No | ||
| 70 | LTA | 23003 | 9883 | -0.005 | -0.3378 | No | ||
| 71 | UBXD3 | 15701 | 10016 | -0.007 | -0.3448 | No | ||
| 72 | ATP1B4 | 12586 | 10310 | -0.010 | -0.3606 | No | ||
| 73 | NIN | 9463 2103 | 10679 | -0.015 | -0.3803 | No | ||
| 74 | IRS4 | 9183 4926 | 10847 | -0.018 | -0.3892 | No | ||
| 75 | ITGA3 | 20284 | 10930 | -0.019 | -0.3934 | No | ||
| 76 | NR4A3 | 9473 16212 5183 | 11250 | -0.023 | -0.4104 | No | ||
| 77 | RGMA | 18213 | 11681 | -0.031 | -0.4334 | No | ||
| 78 | TJP1 | 17803 | 11711 | -0.031 | -0.4346 | No | ||
| 79 | PRDM1 | 19775 3337 | 11762 | -0.032 | -0.4370 | No | ||
| 80 | NFIA | 16172 5170 | 11870 | -0.034 | -0.4425 | No | ||
| 81 | ADAM23 | 10699 4057 | 11893 | -0.034 | -0.4433 | No | ||
| 82 | OTX2 | 9519 | 11917 | -0.035 | -0.4442 | No | ||
| 83 | GRIK2 | 3318 | 11926 | -0.035 | -0.4443 | No | ||
| 84 | REST | 16818 | 11942 | -0.035 | -0.4447 | No | ||
| 85 | C1QTNF3 | 2224 22505 | 11999 | -0.036 | -0.4474 | No | ||
| 86 | HOXA11 | 17146 | 12543 | -0.049 | -0.4763 | No | ||
| 87 | HOXB8 | 9111 | 12628 | -0.051 | -0.4803 | No | ||
| 88 | TNF | 23004 | 12665 | -0.052 | -0.4817 | No | ||
| 89 | PLAG1 | 7148 15949 12161 | 12683 | -0.053 | -0.4821 | No | ||
| 90 | FOXD3 | 9088 | 12815 | -0.055 | -0.4886 | No | ||
| 91 | TWIST2 | 14185 | 13001 | -0.061 | -0.4980 | No | ||
| 92 | CEBPA | 4514 | 13139 | -0.066 | -0.5048 | No | ||
| 93 | BMP5 | 19376 | 13482 | -0.079 | -0.5225 | No | ||
| 94 | SLITRK6 | 21724 | 13944 | -0.108 | -0.5463 | No | ||
| 95 | DOCK3 | 9920 5537 10751 | 14150 | -0.122 | -0.5562 | No | ||
| 96 | TLE3 | 5767 19416 | 14526 | -0.159 | -0.5748 | No | ||
| 97 | SHOX2 | 1802 5432 | 14711 | -0.185 | -0.5829 | No | ||
| 98 | DMD | 24295 2647 | 14880 | -0.212 | -0.5898 | No | ||
| 99 | FUT8 | 2163 21233 | 14919 | -0.220 | -0.5897 | No | ||
| 100 | SSH3 | 3709 10890 | 15005 | -0.238 | -0.5918 | No | ||
| 101 | CRIM1 | 403 | 15091 | -0.256 | -0.5939 | No | ||
| 102 | RPS6KB1 | 7815 1207 13040 | 15117 | -0.261 | -0.5926 | No | ||
| 103 | ETV1 | 4688 8925 8924 | 15380 | -0.333 | -0.6034 | No | ||
| 104 | BRUNOL4 | 2010 4229 | 15546 | -0.403 | -0.6082 | No | ||
| 105 | MAP2K5 | 19088 | 15555 | -0.405 | -0.6045 | No | ||
| 106 | UBR1 | 5828 | 15572 | -0.412 | -0.6012 | No | ||
| 107 | CPNE1 | 11186 | 15613 | -0.426 | -0.5991 | No | ||
| 108 | EGR2 | 8886 | 16019 | -0.677 | -0.6141 | Yes | ||
| 109 | MCC | 552 | 16064 | -0.708 | -0.6093 | Yes | ||
| 110 | NEDD4 | 19384 3101 | 16246 | -0.907 | -0.6099 | Yes | ||
| 111 | ICK | 7135 12146 | 16282 | -0.949 | -0.6022 | Yes | ||
| 112 | BUB3 | 18045 | 16662 | -1.322 | -0.6093 | Yes | ||
| 113 | CAB39L | 21992 | 17036 | -1.838 | -0.6109 | Yes | ||
| 114 | RAB3IP | 19623 | 17220 | -2.176 | -0.5988 | Yes | ||
| 115 | CSNK1E | 6570 2211 11332 | 17264 | -2.241 | -0.5784 | Yes | ||
| 116 | PRKAG1 | 22135 | 17374 | -2.424 | -0.5597 | Yes | ||
| 117 | PAIP2 | 12593 | 17398 | -2.482 | -0.5358 | Yes | ||
| 118 | TOB1 | 20703 | 17438 | -2.536 | -0.5123 | Yes | ||
| 119 | ZHX2 | 11842 | 18038 | -4.076 | -0.5034 | Yes | ||
| 120 | ETV5 | 22630 | 18044 | -4.102 | -0.4621 | Yes | ||
| 121 | MSI2 | 8030 1268 | 18136 | -4.503 | -0.4214 | Yes | ||
| 122 | KLF7 | 8205 | 18211 | -4.915 | -0.3757 | Yes | ||
| 123 | AP1GBP1 | 20725 | 18350 | -5.626 | -0.3262 | Yes | ||
| 124 | FLI1 | 4729 | 18402 | -6.008 | -0.2681 | Yes | ||
| 125 | DUSP1 | 23061 | 18438 | -6.362 | -0.2056 | Yes | ||
| 126 | CDC42EP3 | 6436 | 18482 | -6.904 | -0.1380 | Yes | ||
| 127 | GFRA1 | 9015 | 18490 | -6.979 | -0.0677 | Yes | ||
| 128 | SGK | 9809 3419 | 18514 | -7.351 | 0.0055 | Yes |