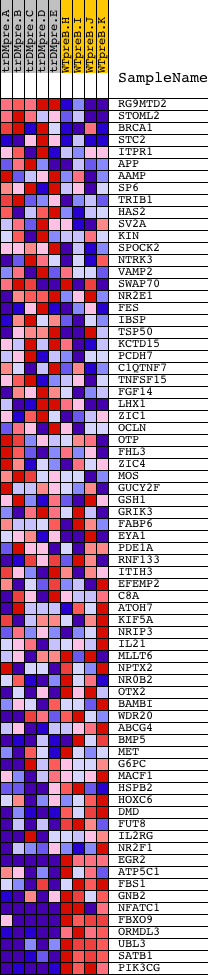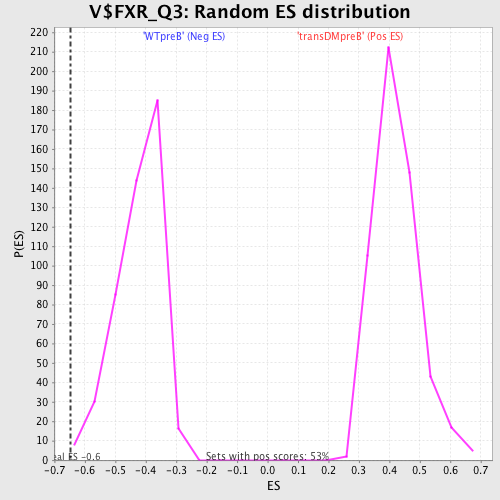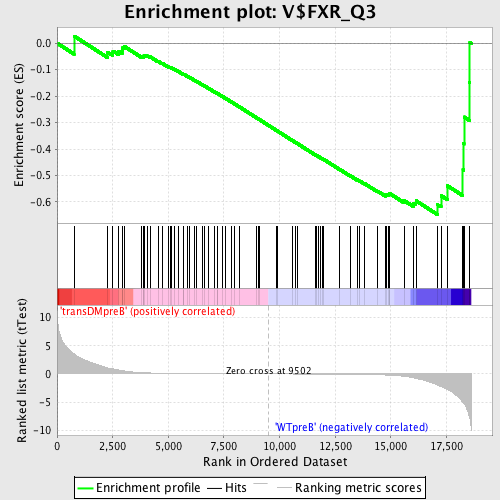
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | V$FXR_Q3 |
| Enrichment Score (ES) | -0.64717716 |
| Normalized Enrichment Score (NES) | -1.535242 |
| Nominal p-value | 0.0064102565 |
| FDR q-value | 0.07961154 |
| FWER p-Value | 0.624 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RG9MTD2 | 8443 | 764 | 3.538 | 0.0267 | No | ||
| 2 | STOML2 | 15902 | 2264 | 1.073 | -0.0335 | No | ||
| 3 | BRCA1 | 20213 | 2509 | 0.891 | -0.0295 | No | ||
| 4 | STC2 | 5526 | 2770 | 0.685 | -0.0304 | No | ||
| 5 | ITPR1 | 17341 | 2923 | 0.571 | -0.0276 | No | ||
| 6 | APP | 4402 | 2943 | 0.560 | -0.0179 | No | ||
| 7 | AAMP | 10406 | 3013 | 0.512 | -0.0118 | No | ||
| 8 | SP6 | 8178 13678 13679 | 3799 | 0.228 | -0.0497 | No | ||
| 9 | TRIB1 | 22467 | 3893 | 0.212 | -0.0507 | No | ||
| 10 | HAS2 | 22293 | 3904 | 0.209 | -0.0472 | No | ||
| 11 | SV2A | 15498 12250 | 3933 | 0.204 | -0.0448 | No | ||
| 12 | KIN | 15120 | 4062 | 0.181 | -0.0482 | No | ||
| 13 | SPOCK2 | 13585 | 4181 | 0.166 | -0.0514 | No | ||
| 14 | NTRK3 | 2069 3652 3705 9492 | 4537 | 0.128 | -0.0681 | No | ||
| 15 | VAMP2 | 20826 | 4730 | 0.114 | -0.0762 | No | ||
| 16 | SWAP70 | 18126 5553 9948 | 5007 | 0.096 | -0.0893 | No | ||
| 17 | NR2E1 | 19780 | 5080 | 0.092 | -0.0914 | No | ||
| 18 | FES | 17779 3949 | 5137 | 0.089 | -0.0927 | No | ||
| 19 | IBSP | 16775 | 5269 | 0.082 | -0.0982 | No | ||
| 20 | TSP50 | 19286 | 5465 | 0.074 | -0.1073 | No | ||
| 21 | KCTD15 | 17864 | 5681 | 0.065 | -0.1176 | No | ||
| 22 | PCDH7 | 7029 3460 | 5693 | 0.065 | -0.1170 | No | ||
| 23 | C1QTNF7 | 16852 | 5866 | 0.060 | -0.1251 | No | ||
| 24 | TNFSF15 | 15863 | 5968 | 0.057 | -0.1295 | No | ||
| 25 | FGF14 | 21716 3005 3463 | 6182 | 0.051 | -0.1400 | No | ||
| 26 | LHX1 | 4995 | 6286 | 0.049 | -0.1446 | No | ||
| 27 | ZIC1 | 19040 | 6531 | 0.044 | -0.1569 | No | ||
| 28 | OCLN | 21368 | 6614 | 0.042 | -0.1605 | No | ||
| 29 | OTP | 21582 | 6826 | 0.038 | -0.1712 | No | ||
| 30 | FHL3 | 16092 2457 | 7082 | 0.033 | -0.1843 | No | ||
| 31 | ZIC4 | 5992 | 7197 | 0.031 | -0.1898 | No | ||
| 32 | MOS | 16254 | 7227 | 0.031 | -0.1908 | No | ||
| 33 | GUCY2F | 24045 | 7454 | 0.027 | -0.2025 | No | ||
| 34 | GSH1 | 16622 | 7569 | 0.025 | -0.2081 | No | ||
| 35 | GRIK3 | 16083 | 7855 | 0.022 | -0.2231 | No | ||
| 36 | FABP6 | 9178 | 7952 | 0.020 | -0.2279 | No | ||
| 37 | EYA1 | 4695 4061 | 8206 | 0.017 | -0.2412 | No | ||
| 38 | PDE1A | 9541 5234 | 8950 | 0.007 | -0.2811 | No | ||
| 39 | RNF133 | 11813 | 9055 | 0.006 | -0.2866 | No | ||
| 40 | ITIH3 | 21896 | 9082 | 0.005 | -0.2879 | No | ||
| 41 | EFEMP2 | 23781 | 9871 | -0.005 | -0.3303 | No | ||
| 42 | C8A | 15823 | 9924 | -0.006 | -0.3330 | No | ||
| 43 | ATOH7 | 20000 | 10599 | -0.014 | -0.3691 | No | ||
| 44 | KIF5A | 9222 | 10720 | -0.016 | -0.3753 | No | ||
| 45 | NRIP3 | 17679 | 10791 | -0.017 | -0.3787 | No | ||
| 46 | IL21 | 12241 | 11629 | -0.029 | -0.4233 | No | ||
| 47 | MLLT6 | 20678 | 11644 | -0.030 | -0.4235 | No | ||
| 48 | NPTX2 | 16630 | 11767 | -0.032 | -0.4294 | No | ||
| 49 | NR0B2 | 16050 | 11856 | -0.033 | -0.4335 | No | ||
| 50 | OTX2 | 9519 | 11917 | -0.035 | -0.4361 | No | ||
| 51 | BAMBI | 23629 | 11965 | -0.035 | -0.4380 | No | ||
| 52 | WDR20 | 21258 | 12709 | -0.053 | -0.4770 | No | ||
| 53 | ABCG4 | 19151 | 13174 | -0.067 | -0.5007 | No | ||
| 54 | BMP5 | 19376 | 13482 | -0.079 | -0.5158 | No | ||
| 55 | MET | 17520 | 13610 | -0.086 | -0.5210 | No | ||
| 56 | G6PC | 20656 | 13820 | -0.099 | -0.5303 | No | ||
| 57 | MACF1 | 4316 15763 2382 15764 | 14400 | -0.145 | -0.5588 | No | ||
| 58 | HSPB2 | 19121 | 14755 | -0.191 | -0.5742 | No | ||
| 59 | HOXC6 | 22339 2302 | 14803 | -0.199 | -0.5729 | No | ||
| 60 | DMD | 24295 2647 | 14880 | -0.212 | -0.5729 | No | ||
| 61 | FUT8 | 2163 21233 | 14919 | -0.220 | -0.5708 | No | ||
| 62 | IL2RG | 24096 | 14951 | -0.226 | -0.5681 | No | ||
| 63 | NR2F1 | 7015 21401 | 15612 | -0.426 | -0.5955 | No | ||
| 64 | EGR2 | 8886 | 16019 | -0.677 | -0.6044 | No | ||
| 65 | ATP5C1 | 8635 | 16131 | -0.780 | -0.5954 | No | ||
| 66 | FBS1 | 18067 | 17092 | -1.944 | -0.6098 | Yes | ||
| 67 | GNB2 | 9026 | 17260 | -2.233 | -0.5760 | Yes | ||
| 68 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 17547 | -2.734 | -0.5389 | Yes | ||
| 69 | FBXO9 | 19058 3146 | 18243 | -5.084 | -0.4787 | Yes | ||
| 70 | ORMDL3 | 12386 | 18267 | -5.225 | -0.3796 | Yes | ||
| 71 | UBL3 | 16284 | 18292 | -5.319 | -0.2787 | Yes | ||
| 72 | SATB1 | 5408 | 18527 | -7.452 | -0.1482 | Yes | ||
| 73 | PIK3CG | 6635 | 18554 | -7.967 | 0.0033 | Yes |