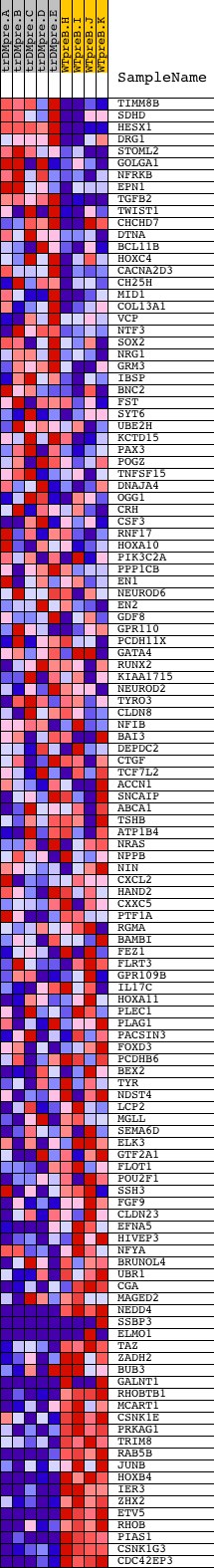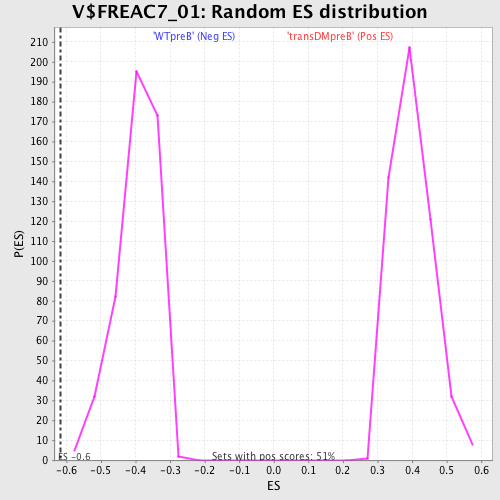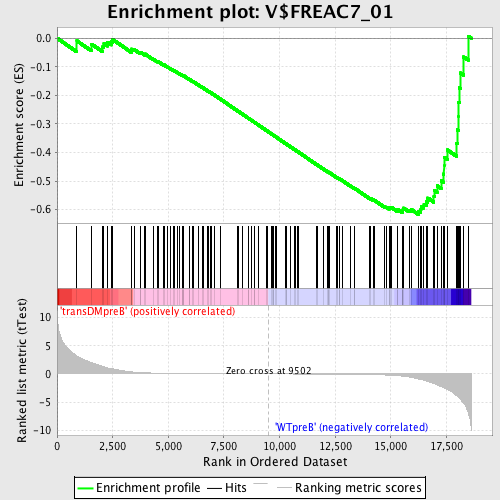
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | V$FREAC7_01 |
| Enrichment Score (ES) | -0.61695147 |
| Normalized Enrichment Score (NES) | -1.5650164 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07489662 |
| FWER p-Value | 0.449 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TIMM8B | 11391 | 890 | 3.180 | -0.0094 | No | ||
| 2 | SDHD | 19125 | 1545 | 1.990 | -0.0206 | No | ||
| 3 | HESX1 | 22070 | 2036 | 1.352 | -0.0306 | No | ||
| 4 | DRG1 | 1199 20553 | 2105 | 1.261 | -0.0190 | No | ||
| 5 | STOML2 | 15902 | 2264 | 1.073 | -0.0144 | No | ||
| 6 | GOLGA1 | 14595 | 2439 | 0.947 | -0.0123 | No | ||
| 7 | NFRKB | 10642 | 2480 | 0.910 | -0.0034 | No | ||
| 8 | EPN1 | 18401 | 3336 | 0.359 | -0.0453 | No | ||
| 9 | TGFB2 | 5743 | 3353 | 0.350 | -0.0419 | No | ||
| 10 | TWIST1 | 21291 | 3357 | 0.347 | -0.0378 | No | ||
| 11 | CHCHD7 | 16280 | 3470 | 0.308 | -0.0401 | No | ||
| 12 | DTNA | 8870 4647 23611 1991 | 3730 | 0.242 | -0.0512 | No | ||
| 13 | BCL11B | 7190 | 3740 | 0.240 | -0.0488 | No | ||
| 14 | HOXC4 | 4864 | 3935 | 0.203 | -0.0568 | No | ||
| 15 | CACNA2D3 | 8677 | 3953 | 0.201 | -0.0553 | No | ||
| 16 | CH25H | 23695 | 4340 | 0.147 | -0.0743 | No | ||
| 17 | MID1 | 5097 5098 | 4512 | 0.131 | -0.0820 | No | ||
| 18 | COL13A1 | 8763 | 4555 | 0.127 | -0.0827 | No | ||
| 19 | VCP | 11254 | 4780 | 0.110 | -0.0935 | No | ||
| 20 | NTF3 | 16991 | 4805 | 0.109 | -0.0935 | No | ||
| 21 | SOX2 | 9849 15612 | 4978 | 0.098 | -0.1016 | No | ||
| 22 | NRG1 | 3885 | 5099 | 0.090 | -0.1070 | No | ||
| 23 | GRM3 | 16929 | 5231 | 0.084 | -0.1130 | No | ||
| 24 | IBSP | 16775 | 5269 | 0.082 | -0.1140 | No | ||
| 25 | BNC2 | 15850 | 5408 | 0.076 | -0.1205 | No | ||
| 26 | FST | 8985 | 5497 | 0.072 | -0.1244 | No | ||
| 27 | SYT6 | 13437 7042 12040 | 5616 | 0.067 | -0.1300 | No | ||
| 28 | UBE2H | 5823 5822 | 5656 | 0.066 | -0.1313 | No | ||
| 29 | KCTD15 | 17864 | 5681 | 0.065 | -0.1318 | No | ||
| 30 | PAX3 | 13910 | 5683 | 0.065 | -0.1311 | No | ||
| 31 | POGZ | 6031 1911 | 5931 | 0.058 | -0.1437 | No | ||
| 32 | TNFSF15 | 15863 | 5968 | 0.057 | -0.1450 | No | ||
| 33 | DNAJA4 | 19446 | 6072 | 0.054 | -0.1499 | No | ||
| 34 | OGG1 | 17331 | 6130 | 0.053 | -0.1523 | No | ||
| 35 | CRH | 8784 | 6337 | 0.047 | -0.1629 | No | ||
| 36 | CSF3 | 1394 20671 | 6539 | 0.043 | -0.1732 | No | ||
| 37 | RNF17 | 11389 | 6557 | 0.043 | -0.1736 | No | ||
| 38 | HOXA10 | 17145 989 | 6757 | 0.039 | -0.1839 | No | ||
| 39 | PIK3C2A | 17667 | 6785 | 0.038 | -0.1849 | No | ||
| 40 | PPP1CB | 5282 3529 3504 | 6889 | 0.037 | -0.1900 | No | ||
| 41 | EN1 | 387 14155 | 6942 | 0.036 | -0.1924 | No | ||
| 42 | NEUROD6 | 17138 | 7093 | 0.033 | -0.2001 | No | ||
| 43 | EN2 | 16898 | 7357 | 0.028 | -0.2140 | No | ||
| 44 | GDF8 | 9411 | 8129 | 0.018 | -0.2554 | No | ||
| 45 | GPR110 | 23234 | 8163 | 0.017 | -0.2570 | No | ||
| 46 | PCDH11X | 2559 | 8348 | 0.015 | -0.2668 | No | ||
| 47 | GATA4 | 21792 4755 | 8608 | 0.011 | -0.2807 | No | ||
| 48 | RUNX2 | 4480 8700 | 8624 | 0.011 | -0.2813 | No | ||
| 49 | KIAA1715 | 7629 | 8738 | 0.010 | -0.2873 | No | ||
| 50 | NEUROD2 | 20262 | 8856 | 0.008 | -0.2936 | No | ||
| 51 | TYRO3 | 5811 | 8858 | 0.008 | -0.2935 | No | ||
| 52 | CLDN8 | 22549 1747 | 9068 | 0.006 | -0.3047 | No | ||
| 53 | NFIB | 15855 | 9395 | 0.001 | -0.3224 | No | ||
| 54 | BAI3 | 5580 | 9437 | 0.001 | -0.3246 | No | ||
| 55 | DEPDC2 | 14295 | 9655 | -0.002 | -0.3363 | No | ||
| 56 | CTGF | 20068 | 9676 | -0.002 | -0.3373 | No | ||
| 57 | TCF7L2 | 10048 5646 | 9716 | -0.003 | -0.3394 | No | ||
| 58 | ACCN1 | 20327 1194 | 9718 | -0.003 | -0.3394 | No | ||
| 59 | SNCAIP | 12589 | 9831 | -0.005 | -0.3454 | No | ||
| 60 | ABCA1 | 15881 | 9838 | -0.005 | -0.3457 | No | ||
| 61 | TSHB | 15218 | 10280 | -0.010 | -0.3694 | No | ||
| 62 | ATP1B4 | 12586 | 10310 | -0.010 | -0.3708 | No | ||
| 63 | NRAS | 5191 | 10314 | -0.010 | -0.3709 | No | ||
| 64 | NPPB | 15994 | 10485 | -0.013 | -0.3799 | No | ||
| 65 | NIN | 9463 2103 | 10679 | -0.015 | -0.3902 | No | ||
| 66 | CXCL2 | 9790 | 10702 | -0.016 | -0.3912 | No | ||
| 67 | HAND2 | 18615 | 10785 | -0.017 | -0.3954 | No | ||
| 68 | CXXC5 | 12523 | 10860 | -0.018 | -0.3992 | No | ||
| 69 | PTF1A | 15110 | 11670 | -0.030 | -0.4426 | No | ||
| 70 | RGMA | 18213 | 11681 | -0.031 | -0.4427 | No | ||
| 71 | BAMBI | 23629 | 11965 | -0.035 | -0.4576 | No | ||
| 72 | FEZ1 | 19520 3100 | 12153 | -0.039 | -0.4672 | No | ||
| 73 | FLRT3 | 7730 12930 2673 | 12180 | -0.039 | -0.4682 | No | ||
| 74 | GPR109B | 16374 | 12213 | -0.040 | -0.4694 | No | ||
| 75 | IL17C | 18438 | 12261 | -0.042 | -0.4714 | No | ||
| 76 | HOXA11 | 17146 | 12543 | -0.049 | -0.4860 | No | ||
| 77 | PLEC1 | 2273 2198 2205 2178 2230 22247 2213 2231 2295 2172 5263 2266 | 12586 | -0.050 | -0.4877 | No | ||
| 78 | PLAG1 | 7148 15949 12161 | 12683 | -0.053 | -0.4923 | No | ||
| 79 | PACSIN3 | 14948 8149 2776 2665 | 12704 | -0.053 | -0.4927 | No | ||
| 80 | FOXD3 | 9088 | 12815 | -0.055 | -0.4980 | No | ||
| 81 | PCDHB6 | 8224 | 13191 | -0.068 | -0.5174 | No | ||
| 82 | BEX2 | 24055 | 13349 | -0.074 | -0.5250 | No | ||
| 83 | TYR | 17763 | 13354 | -0.074 | -0.5243 | No | ||
| 84 | NDST4 | 12259 7228 | 14040 | -0.114 | -0.5600 | No | ||
| 85 | LCP2 | 4988 9268 | 14104 | -0.118 | -0.5619 | No | ||
| 86 | MGLL | 17368 1145 | 14199 | -0.127 | -0.5655 | No | ||
| 87 | SEMA6D | 2800 14876 90 | 14269 | -0.133 | -0.5676 | No | ||
| 88 | ELK3 | 4663 94 3388 | 14736 | -0.189 | -0.5905 | No | ||
| 89 | GTF2A1 | 2064 8187 8188 13512 | 14800 | -0.198 | -0.5915 | No | ||
| 90 | FLOT1 | 23249 1613 | 14922 | -0.221 | -0.5954 | No | ||
| 91 | POU2F1 | 5275 3989 4065 4010 | 14949 | -0.225 | -0.5940 | No | ||
| 92 | SSH3 | 3709 10890 | 15005 | -0.238 | -0.5941 | No | ||
| 93 | FGF9 | 8966 | 15041 | -0.246 | -0.5930 | No | ||
| 94 | CLDN23 | 18893 | 15315 | -0.313 | -0.6040 | No | ||
| 95 | EFNA5 | 4655 8884 | 15319 | -0.314 | -0.6003 | No | ||
| 96 | HIVEP3 | 4973 | 15528 | -0.393 | -0.6068 | Yes | ||
| 97 | NFYA | 5172 | 15535 | -0.397 | -0.6022 | Yes | ||
| 98 | BRUNOL4 | 2010 4229 | 15546 | -0.403 | -0.5979 | Yes | ||
| 99 | UBR1 | 5828 | 15572 | -0.412 | -0.5942 | Yes | ||
| 100 | CGA | 16246 | 15826 | -0.528 | -0.6015 | Yes | ||
| 101 | MAGED2 | 24034 | 15912 | -0.585 | -0.5989 | Yes | ||
| 102 | NEDD4 | 19384 3101 | 16246 | -0.907 | -0.6059 | Yes | ||
| 103 | SSBP3 | 2439 7814 2431 | 16349 | -0.999 | -0.5993 | Yes | ||
| 104 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 16358 | -1.001 | -0.5875 | Yes | ||
| 105 | TAZ | 12415 | 16490 | -1.117 | -0.5810 | Yes | ||
| 106 | ZADH2 | 23501 | 16598 | -1.223 | -0.5719 | Yes | ||
| 107 | BUB3 | 18045 | 16662 | -1.322 | -0.5592 | Yes | ||
| 108 | GALNT1 | 8998 23609 8999 4750 | 16937 | -1.721 | -0.5531 | Yes | ||
| 109 | RHOBTB1 | 12789 | 16963 | -1.732 | -0.5334 | Yes | ||
| 110 | MCART1 | 10492 | 17083 | -1.929 | -0.5164 | Yes | ||
| 111 | CSNK1E | 6570 2211 11332 | 17264 | -2.241 | -0.4988 | Yes | ||
| 112 | PRKAG1 | 22135 | 17374 | -2.424 | -0.4752 | Yes | ||
| 113 | TRIM8 | 23824 | 17390 | -2.463 | -0.4460 | Yes | ||
| 114 | RAB5B | 359 19591 3430 | 17395 | -2.480 | -0.4161 | Yes | ||
| 115 | JUNB | 9201 | 17543 | -2.722 | -0.3909 | Yes | ||
| 116 | HOXB4 | 20686 | 17960 | -3.831 | -0.3668 | Yes | ||
| 117 | IER3 | 23248 | 17998 | -3.945 | -0.3208 | Yes | ||
| 118 | ZHX2 | 11842 | 18038 | -4.076 | -0.2733 | Yes | ||
| 119 | ETV5 | 22630 | 18044 | -4.102 | -0.2237 | Yes | ||
| 120 | RHOB | 4411 | 18087 | -4.288 | -0.1738 | Yes | ||
| 121 | PIAS1 | 7126 | 18126 | -4.458 | -0.1216 | Yes | ||
| 122 | CSNK1G3 | 12873 | 18274 | -5.253 | -0.0656 | Yes | ||
| 123 | CDC42EP3 | 6436 | 18482 | -6.904 | 0.0072 | Yes |