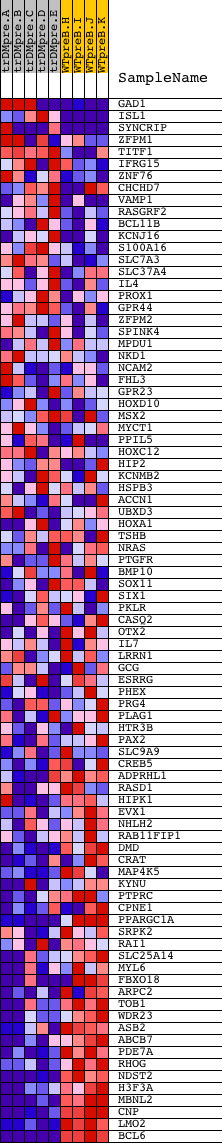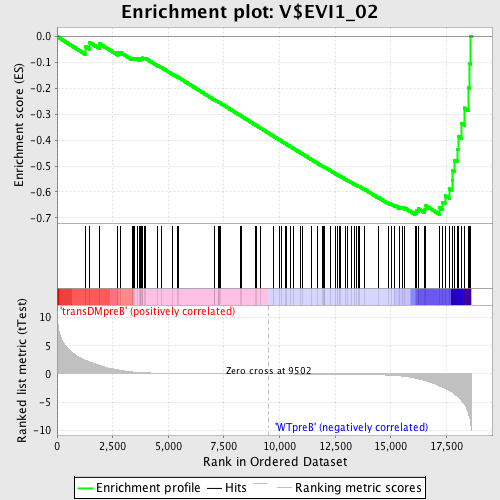
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | V$EVI1_02 |
| Enrichment Score (ES) | -0.68686527 |
| Normalized Enrichment Score (NES) | -1.6370817 |
| Nominal p-value | 0.0020833334 |
| FDR q-value | 0.056103114 |
| FWER p-Value | 0.163 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GAD1 | 14993 2690 | 1262 | 2.444 | -0.0380 | No | ||
| 2 | ISL1 | 21338 | 1455 | 2.133 | -0.0220 | No | ||
| 3 | SYNCRIP | 3078 3035 3107 | 1899 | 1.539 | -0.0269 | No | ||
| 4 | ZFPM1 | 18439 | 2724 | 0.723 | -0.0625 | No | ||
| 5 | TITF1 | 10180 21062 | 2856 | 0.623 | -0.0619 | No | ||
| 6 | IFRG15 | 7222 | 3376 | 0.339 | -0.0857 | No | ||
| 7 | ZNF76 | 23320 | 3416 | 0.325 | -0.0838 | No | ||
| 8 | CHCHD7 | 16280 | 3470 | 0.308 | -0.0829 | No | ||
| 9 | VAMP1 | 5846 1043 | 3597 | 0.273 | -0.0863 | No | ||
| 10 | RASGRF2 | 21397 | 3704 | 0.248 | -0.0890 | No | ||
| 11 | BCL11B | 7190 | 3740 | 0.240 | -0.0879 | No | ||
| 12 | KCNJ16 | 20613 | 3757 | 0.236 | -0.0858 | No | ||
| 13 | S100A16 | 15530 | 3775 | 0.232 | -0.0839 | No | ||
| 14 | SLC7A3 | 2608 24097 | 3845 | 0.219 | -0.0849 | No | ||
| 15 | SLC37A4 | 8994 | 3853 | 0.218 | -0.0826 | No | ||
| 16 | IL4 | 9174 | 3912 | 0.208 | -0.0832 | No | ||
| 17 | PROX1 | 9623 | 3960 | 0.200 | -0.0832 | No | ||
| 18 | GPR44 | 23926 | 4533 | 0.129 | -0.1125 | No | ||
| 19 | ZFPM2 | 22483 | 4689 | 0.116 | -0.1195 | No | ||
| 20 | SPINK4 | 16241 | 5206 | 0.085 | -0.1463 | No | ||
| 21 | MPDU1 | 20386 | 5389 | 0.077 | -0.1551 | No | ||
| 22 | NKD1 | 8228 | 5466 | 0.074 | -0.1583 | No | ||
| 23 | NCAM2 | 9445 5150 | 7062 | 0.034 | -0.2440 | No | ||
| 24 | FHL3 | 16092 2457 | 7082 | 0.033 | -0.2446 | No | ||
| 25 | GPR23 | 13428 | 7235 | 0.031 | -0.2524 | No | ||
| 26 | HOXD10 | 14983 | 7258 | 0.030 | -0.2532 | No | ||
| 27 | MSX2 | 21461 | 7300 | 0.030 | -0.2551 | No | ||
| 28 | MYCT1 | 7572 | 7333 | 0.029 | -0.2565 | No | ||
| 29 | PPIL5 | 21255 | 8250 | 0.016 | -0.3057 | No | ||
| 30 | HOXC12 | 22343 | 8308 | 0.015 | -0.3086 | No | ||
| 31 | HIP2 | 3626 3485 16838 | 8926 | 0.007 | -0.3418 | No | ||
| 32 | KCNMB2 | 1939 15615 | 8955 | 0.007 | -0.3432 | No | ||
| 33 | HSPB3 | 21340 | 9153 | 0.005 | -0.3538 | No | ||
| 34 | ACCN1 | 20327 1194 | 9718 | -0.003 | -0.3842 | No | ||
| 35 | UBXD3 | 15701 | 10016 | -0.007 | -0.4001 | No | ||
| 36 | HOXA1 | 1005 17152 | 10104 | -0.008 | -0.4047 | No | ||
| 37 | TSHB | 15218 | 10280 | -0.010 | -0.4141 | No | ||
| 38 | NRAS | 5191 | 10314 | -0.010 | -0.4157 | No | ||
| 39 | PTGFR | 15138 | 10503 | -0.013 | -0.4257 | No | ||
| 40 | BMP10 | 17376 | 10617 | -0.014 | -0.4316 | No | ||
| 41 | SOX11 | 5477 | 10943 | -0.019 | -0.4489 | No | ||
| 42 | SIX1 | 9821 | 11012 | -0.020 | -0.4523 | No | ||
| 43 | PKLR | 1850 15545 | 11428 | -0.026 | -0.4744 | No | ||
| 44 | CASQ2 | 15477 | 11687 | -0.031 | -0.4880 | No | ||
| 45 | OTX2 | 9519 | 11917 | -0.035 | -0.4999 | No | ||
| 46 | IL7 | 4921 | 11984 | -0.036 | -0.5030 | No | ||
| 47 | LRRN1 | 17343 | 12010 | -0.036 | -0.5039 | No | ||
| 48 | GCG | 14578 | 12019 | -0.036 | -0.5039 | No | ||
| 49 | ESRRG | 6447 | 12274 | -0.042 | -0.5171 | No | ||
| 50 | PHEX | 24024 | 12530 | -0.049 | -0.5303 | No | ||
| 51 | PRG4 | 13592 4036 | 12604 | -0.050 | -0.5336 | No | ||
| 52 | PLAG1 | 7148 15949 12161 | 12683 | -0.053 | -0.5371 | No | ||
| 53 | HTR3B | 19129 3145 | 12730 | -0.053 | -0.5390 | No | ||
| 54 | PAX2 | 9530 24416 | 12961 | -0.060 | -0.5506 | No | ||
| 55 | SLC9A9 | 11661 | 13046 | -0.063 | -0.5544 | No | ||
| 56 | CREB5 | 10551 | 13238 | -0.070 | -0.5638 | No | ||
| 57 | ADPRHL1 | 18930 3863 | 13353 | -0.074 | -0.5691 | No | ||
| 58 | RASD1 | 20424 | 13475 | -0.079 | -0.5746 | No | ||
| 59 | HIPK1 | 4851 | 13544 | -0.082 | -0.5773 | No | ||
| 60 | EVX1 | 17442 | 13581 | -0.084 | -0.5782 | No | ||
| 61 | NHLH2 | 9462 | 13811 | -0.098 | -0.5893 | No | ||
| 62 | RAB11FIP1 | 13287 | 14461 | -0.152 | -0.6225 | No | ||
| 63 | DMD | 24295 2647 | 14880 | -0.212 | -0.6424 | No | ||
| 64 | CRAT | 4557 2804 2896 | 15007 | -0.238 | -0.6463 | No | ||
| 65 | MAP4K5 | 11853 21048 2101 2142 | 15180 | -0.275 | -0.6522 | No | ||
| 66 | KYNU | 15016 | 15411 | -0.345 | -0.6603 | No | ||
| 67 | PTPRC | 5327 9662 | 15502 | -0.380 | -0.6605 | No | ||
| 68 | CPNE1 | 11186 | 15613 | -0.426 | -0.6612 | No | ||
| 69 | PPARGC1A | 16533 | 16090 | -0.735 | -0.6778 | Yes | ||
| 70 | SRPK2 | 5513 | 16172 | -0.834 | -0.6719 | Yes | ||
| 71 | RAI1 | 5355 20861 | 16248 | -0.910 | -0.6647 | Yes | ||
| 72 | SLC25A14 | 24344 | 16533 | -1.153 | -0.6658 | Yes | ||
| 73 | MYL6 | 9438 3408 | 16575 | -1.199 | -0.6532 | Yes | ||
| 74 | FBXO18 | 14686 2796 | 17172 | -2.092 | -0.6596 | Yes | ||
| 75 | ARPC2 | 14225 | 17323 | -2.337 | -0.6389 | Yes | ||
| 76 | TOB1 | 20703 | 17438 | -2.536 | -0.6137 | Yes | ||
| 77 | WDR23 | 22009 | 17625 | -2.889 | -0.5881 | Yes | ||
| 78 | ASB2 | 20996 | 17751 | -3.188 | -0.5556 | Yes | ||
| 79 | ABCB7 | 24085 | 17789 | -3.335 | -0.5164 | Yes | ||
| 80 | PDE7A | 9544 1788 | 17870 | -3.580 | -0.4766 | Yes | ||
| 81 | RHOG | 17727 | 18003 | -3.959 | -0.4349 | Yes | ||
| 82 | NDST2 | 21906 | 18058 | -4.144 | -0.3867 | Yes | ||
| 83 | H3F3A | 4838 | 18169 | -4.686 | -0.3348 | Yes | ||
| 84 | MBNL2 | 21932 | 18308 | -5.400 | -0.2757 | Yes | ||
| 85 | CNP | 8760 | 18502 | -7.122 | -0.1982 | Yes | ||
| 86 | LMO2 | 9281 | 18546 | -7.809 | -0.1042 | Yes | ||
| 87 | BCL6 | 22624 | 18584 | -8.751 | 0.0017 | Yes |