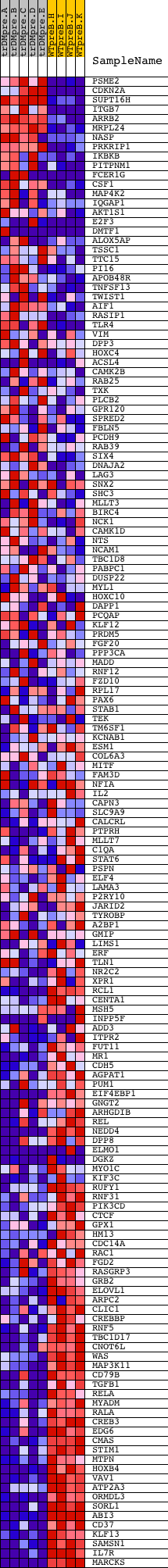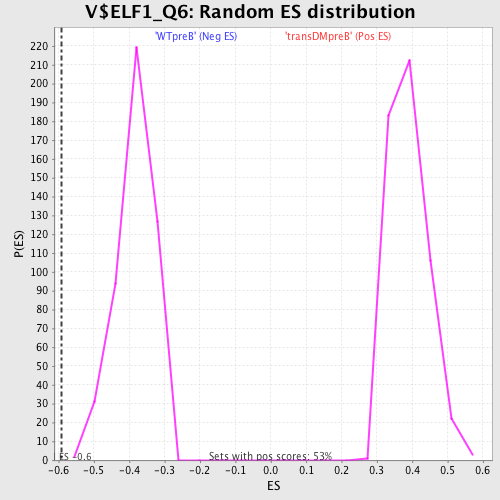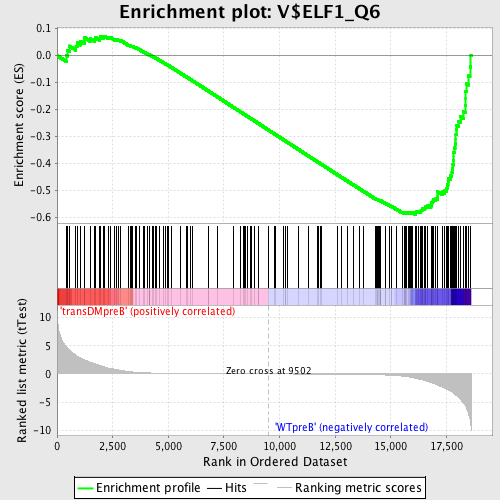
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | V$ELF1_Q6 |
| Enrichment Score (ES) | -0.590264 |
| Normalized Enrichment Score (NES) | -1.5381923 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.084463805 |
| FWER p-Value | 0.611 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSME2 | 5301 | 409 | 4.848 | 0.0010 | No | ||
| 2 | CDKN2A | 2491 15841 | 473 | 4.544 | 0.0194 | No | ||
| 3 | SUPT16H | 8541 | 552 | 4.202 | 0.0353 | No | ||
| 4 | ITGB7 | 22112 | 848 | 3.298 | 0.0351 | No | ||
| 5 | ARRB2 | 20806 | 899 | 3.154 | 0.0474 | No | ||
| 6 | MRPL24 | 15556 | 1059 | 2.790 | 0.0522 | No | ||
| 7 | NASP | 2383 2387 6955 | 1225 | 2.494 | 0.0552 | No | ||
| 8 | PRKRIP1 | 12407 7336 | 1229 | 2.488 | 0.0669 | No | ||
| 9 | IKBKB | 4907 | 1483 | 2.082 | 0.0632 | No | ||
| 10 | PITPNM1 | 23770 | 1680 | 1.795 | 0.0611 | No | ||
| 11 | FCER1G | 13759 | 1722 | 1.741 | 0.0673 | No | ||
| 12 | CSF1 | 15202 | 1912 | 1.525 | 0.0643 | No | ||
| 13 | MAP4K2 | 6457 | 1945 | 1.471 | 0.0696 | No | ||
| 14 | IQGAP1 | 6619 | 2068 | 1.318 | 0.0693 | No | ||
| 15 | AKT1S1 | 12557 | 2130 | 1.234 | 0.0719 | No | ||
| 16 | E2F3 | 21500 8874 | 2288 | 1.048 | 0.0684 | No | ||
| 17 | DMTF1 | 887 3662 16928 3530 | 2378 | 0.994 | 0.0684 | No | ||
| 18 | ALOX5AP | 16616 | 2587 | 0.835 | 0.0611 | No | ||
| 19 | TSSC1 | 2053 2063 6890 2144 | 2690 | 0.748 | 0.0591 | No | ||
| 20 | TTC15 | 5720 | 2780 | 0.673 | 0.0575 | No | ||
| 21 | PI16 | 23310 | 2870 | 0.611 | 0.0556 | No | ||
| 22 | APOB48R | 18081 | 3217 | 0.403 | 0.0388 | No | ||
| 23 | TNFSF13 | 12806 | 3304 | 0.371 | 0.0359 | No | ||
| 24 | TWIST1 | 21291 | 3357 | 0.347 | 0.0348 | No | ||
| 25 | AIF1 | 1582 23005 | 3382 | 0.337 | 0.0351 | No | ||
| 26 | RASIP1 | 12838 | 3521 | 0.292 | 0.0290 | No | ||
| 27 | TLR4 | 2329 10191 5770 | 3553 | 0.285 | 0.0287 | No | ||
| 28 | VIM | 80 | 3689 | 0.250 | 0.0226 | No | ||
| 29 | DPP3 | 7954 3746 | 3869 | 0.215 | 0.0139 | No | ||
| 30 | HOXC4 | 4864 | 3935 | 0.203 | 0.0114 | No | ||
| 31 | ACSL4 | 24043 11936 24042 2614 6940 | 4058 | 0.183 | 0.0056 | No | ||
| 32 | CAMK2B | 20536 | 4132 | 0.173 | 0.0025 | No | ||
| 33 | RAB25 | 15287 | 4137 | 0.172 | 0.0031 | No | ||
| 34 | TXK | 16513 | 4155 | 0.170 | 0.0030 | No | ||
| 35 | PLCB2 | 5262 | 4288 | 0.153 | -0.0034 | No | ||
| 36 | GPR120 | 23869 | 4329 | 0.149 | -0.0049 | No | ||
| 37 | SPRED2 | 8539 4332 | 4420 | 0.139 | -0.0091 | No | ||
| 38 | FBLN5 | 21002 | 4464 | 0.136 | -0.0108 | No | ||
| 39 | PCDH9 | 21736 | 4618 | 0.122 | -0.0185 | No | ||
| 40 | RAB39 | 19113 | 4790 | 0.110 | -0.0272 | No | ||
| 41 | SIX4 | 5445 | 4856 | 0.106 | -0.0302 | No | ||
| 42 | DNAJA2 | 12133 18806 | 4973 | 0.098 | -0.0360 | No | ||
| 43 | LAG3 | 17000 | 5021 | 0.095 | -0.0381 | No | ||
| 44 | SNX2 | 1974 23554 12585 | 5138 | 0.089 | -0.0440 | No | ||
| 45 | SHC3 | 21465 | 5545 | 0.070 | -0.0656 | No | ||
| 46 | MLLT3 | 2359 15847 2413 | 5803 | 0.061 | -0.0793 | No | ||
| 47 | BIRC4 | 2638 8609 | 5839 | 0.060 | -0.0809 | No | ||
| 48 | NCK1 | 9447 5152 | 5996 | 0.056 | -0.0891 | No | ||
| 49 | CAMK1D | 5977 2948 | 6079 | 0.054 | -0.0933 | No | ||
| 50 | NTS | 19635 3297 | 6804 | 0.038 | -0.1323 | No | ||
| 51 | NCAM1 | 5149 | 6809 | 0.038 | -0.1323 | No | ||
| 52 | TBC1D8 | 12044 | 7192 | 0.031 | -0.1529 | No | ||
| 53 | PABPC1 | 5219 9522 9523 23572 | 7921 | 0.021 | -0.1922 | No | ||
| 54 | DUSP22 | 466 21680 | 8256 | 0.016 | -0.2102 | No | ||
| 55 | MYL1 | 4015 | 8369 | 0.015 | -0.2162 | No | ||
| 56 | HOXC10 | 22342 | 8430 | 0.014 | -0.2194 | No | ||
| 57 | DAPP1 | 11159 6445 6446 | 8472 | 0.013 | -0.2216 | No | ||
| 58 | PCQAP | 1661 13581 | 8575 | 0.012 | -0.2270 | No | ||
| 59 | KLF12 | 4960 2637 21732 9228 | 8710 | 0.010 | -0.2343 | No | ||
| 60 | PRDM5 | 17424 | 8736 | 0.010 | -0.2356 | No | ||
| 61 | FGF20 | 18892 | 8888 | 0.008 | -0.2437 | No | ||
| 62 | PPP3CA | 1863 5284 | 9030 | 0.006 | -0.2513 | No | ||
| 63 | MADD | 2926 14526 | 9048 | 0.006 | -0.2522 | No | ||
| 64 | RNF12 | 5384 | 9510 | -0.000 | -0.2772 | No | ||
| 65 | FZD10 | 13565 16693 | 9787 | -0.004 | -0.2921 | No | ||
| 66 | RPL17 | 11429 6653 | 9805 | -0.004 | -0.2930 | No | ||
| 67 | PAX6 | 5223 | 10181 | -0.009 | -0.3133 | No | ||
| 68 | STAB1 | 21892 | 10249 | -0.010 | -0.3169 | No | ||
| 69 | TEK | 16175 | 10277 | -0.010 | -0.3183 | No | ||
| 70 | TM6SF1 | 8416 | 10362 | -0.011 | -0.3228 | No | ||
| 71 | KCNAB1 | 1791 15577 | 10868 | -0.018 | -0.3501 | No | ||
| 72 | ESM1 | 21560 | 11279 | -0.024 | -0.3722 | No | ||
| 73 | COL6A3 | 4546 13885 13884 | 11713 | -0.031 | -0.3955 | No | ||
| 74 | MITF | 17349 | 11768 | -0.032 | -0.3983 | No | ||
| 75 | FAM3D | 21922 | 11822 | -0.033 | -0.4010 | No | ||
| 76 | NFIA | 16172 5170 | 11870 | -0.034 | -0.4034 | No | ||
| 77 | IL2 | 15354 | 12608 | -0.050 | -0.4430 | No | ||
| 78 | CAPN3 | 8686 2868 | 12787 | -0.055 | -0.4524 | No | ||
| 79 | SLC9A9 | 11661 | 13046 | -0.063 | -0.4661 | No | ||
| 80 | CALCRL | 12041 | 13332 | -0.073 | -0.4812 | No | ||
| 81 | PTPRH | 11861 | 13593 | -0.085 | -0.4949 | No | ||
| 82 | MLLT7 | 24278 | 13790 | -0.097 | -0.5050 | No | ||
| 83 | C1QA | 8666 | 14321 | -0.137 | -0.5331 | No | ||
| 84 | STAT6 | 19854 9909 | 14362 | -0.141 | -0.5346 | No | ||
| 85 | PSPN | 22921 | 14386 | -0.144 | -0.5351 | No | ||
| 86 | ELF4 | 24162 | 14404 | -0.145 | -0.5354 | No | ||
| 87 | LAMA3 | 23620 2009 | 14454 | -0.152 | -0.5373 | No | ||
| 88 | P2RY10 | 2613 24269 | 14484 | -0.155 | -0.5381 | No | ||
| 89 | JARID2 | 9200 | 14516 | -0.158 | -0.5390 | No | ||
| 90 | TYROBP | 18302 | 14544 | -0.162 | -0.5397 | No | ||
| 91 | A2BP1 | 11212 1652 1689 | 14547 | -0.162 | -0.5391 | No | ||
| 92 | GMIP | 18597 | 14765 | -0.193 | -0.5499 | No | ||
| 93 | LIMS1 | 8493 4290 | 14778 | -0.195 | -0.5496 | No | ||
| 94 | ERF | 4680 8914 | 14929 | -0.222 | -0.5567 | No | ||
| 95 | TLN1 | 15899 | 15017 | -0.241 | -0.5602 | No | ||
| 96 | NR2C2 | 10216 | 15039 | -0.245 | -0.5602 | No | ||
| 97 | XPR1 | 5383 | 15248 | -0.294 | -0.5701 | No | ||
| 98 | RCL1 | 3728 3743 23894 | 15515 | -0.386 | -0.5826 | No | ||
| 99 | CENTA1 | 10546 16319 | 15608 | -0.424 | -0.5856 | No | ||
| 100 | MSH5 | 23010 1599 | 15631 | -0.435 | -0.5847 | No | ||
| 101 | INPP5F | 18055 3877 | 15664 | -0.448 | -0.5843 | No | ||
| 102 | ADD3 | 23812 3694 | 15672 | -0.451 | -0.5825 | No | ||
| 103 | ITPR2 | 9194 | 15707 | -0.466 | -0.5821 | No | ||
| 104 | FUT11 | 13084 | 15789 | -0.509 | -0.5841 | No | ||
| 105 | MR1 | 4837 | 15818 | -0.524 | -0.5831 | No | ||
| 106 | CDH5 | 18512 | 15869 | -0.553 | -0.5831 | No | ||
| 107 | AGPAT1 | 23275 | 15921 | -0.594 | -0.5831 | No | ||
| 108 | PUM1 | 8160 | 15982 | -0.646 | -0.5832 | No | ||
| 109 | EIF4EBP1 | 8891 4661 | 16113 | -0.761 | -0.5866 | Yes | ||
| 110 | GNGT2 | 20691 | 16116 | -0.765 | -0.5831 | Yes | ||
| 111 | ARHGDIB | 16950 | 16142 | -0.803 | -0.5806 | Yes | ||
| 112 | REL | 9716 | 16159 | -0.818 | -0.5775 | Yes | ||
| 113 | NEDD4 | 19384 3101 | 16246 | -0.907 | -0.5778 | Yes | ||
| 114 | DPP8 | 19408 | 16324 | -0.985 | -0.5773 | Yes | ||
| 115 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 16358 | -1.001 | -0.5743 | Yes | ||
| 116 | DGKZ | 2836 14522 | 16405 | -1.032 | -0.5719 | Yes | ||
| 117 | MYO1C | 20777 | 16413 | -1.040 | -0.5673 | Yes | ||
| 118 | KIF3C | 9220 4954 | 16514 | -1.135 | -0.5672 | Yes | ||
| 119 | RUFY1 | 20476 | 16554 | -1.174 | -0.5637 | Yes | ||
| 120 | RNF31 | 11206 | 16577 | -1.201 | -0.5592 | Yes | ||
| 121 | PIK3CD | 9563 | 16667 | -1.326 | -0.5577 | Yes | ||
| 122 | CTCF | 18490 | 16806 | -1.517 | -0.5579 | Yes | ||
| 123 | GPX1 | 19310 | 16820 | -1.535 | -0.5512 | Yes | ||
| 124 | HM13 | 2880 2707 14793 | 16834 | -1.558 | -0.5445 | Yes | ||
| 125 | CDC14A | 15184 | 16894 | -1.662 | -0.5397 | Yes | ||
| 126 | RAC1 | 16302 | 16919 | -1.693 | -0.5329 | Yes | ||
| 127 | FGD2 | 23309 | 17027 | -1.827 | -0.5300 | Yes | ||
| 128 | RASGRP3 | 10767 | 17096 | -1.946 | -0.5243 | Yes | ||
| 129 | GRB2 | 20149 | 17099 | -1.953 | -0.5151 | Yes | ||
| 130 | ELOVL1 | 16114 2484 12026 | 17100 | -1.959 | -0.5057 | Yes | ||
| 131 | ARPC2 | 14225 | 17323 | -2.337 | -0.5066 | Yes | ||
| 132 | CLIC1 | 23262 | 17429 | -2.526 | -0.5002 | Yes | ||
| 133 | CREBBP | 22682 8783 | 17516 | -2.659 | -0.4921 | Yes | ||
| 134 | RNF5 | 63 | 17531 | -2.695 | -0.4800 | Yes | ||
| 135 | TBC1D17 | 6121 | 17571 | -2.780 | -0.4688 | Yes | ||
| 136 | CNOT6L | 6084 | 17586 | -2.819 | -0.4560 | Yes | ||
| 137 | WAS | 24193 | 17688 | -3.040 | -0.4470 | Yes | ||
| 138 | MAP3K11 | 11163 | 17746 | -3.174 | -0.4349 | Yes | ||
| 139 | CD79B | 20185 1309 | 17759 | -3.226 | -0.4201 | Yes | ||
| 140 | TGFB1 | 18332 | 17790 | -3.337 | -0.4057 | Yes | ||
| 141 | RELA | 23783 | 17808 | -3.389 | -0.3904 | Yes | ||
| 142 | MYADM | 11946 | 17826 | -3.450 | -0.3749 | Yes | ||
| 143 | RALA | 21541 | 17827 | -3.453 | -0.3583 | Yes | ||
| 144 | CREB3 | 16231 | 17863 | -3.556 | -0.3432 | Yes | ||
| 145 | EDG6 | 19672 | 17886 | -3.639 | -0.3270 | Yes | ||
| 146 | CMAS | 17245 | 17923 | -3.740 | -0.3110 | Yes | ||
| 147 | STIM1 | 18164 | 17926 | -3.744 | -0.2932 | Yes | ||
| 148 | MTPN | 17183 | 17952 | -3.806 | -0.2764 | Yes | ||
| 149 | HOXB4 | 20686 | 17960 | -3.831 | -0.2584 | Yes | ||
| 150 | VAV1 | 23173 | 18053 | -4.129 | -0.2436 | Yes | ||
| 151 | ATP2A3 | 20795 1314 | 18120 | -4.431 | -0.2260 | Yes | ||
| 152 | ORMDL3 | 12386 | 18267 | -5.225 | -0.2089 | Yes | ||
| 153 | SORL1 | 5474 | 18351 | -5.626 | -0.1865 | Yes | ||
| 154 | ABI3 | 7314 | 18366 | -5.724 | -0.1599 | Yes | ||
| 155 | CD37 | 17835 | 18369 | -5.744 | -0.1325 | Yes | ||
| 156 | KLF13 | 17807 | 18416 | -6.159 | -0.1055 | Yes | ||
| 157 | SAMSN1 | 7485 | 18496 | -7.084 | -0.0759 | Yes | ||
| 158 | IL7R | 9175 4922 | 18559 | -8.025 | -0.0408 | Yes | ||
| 159 | MARCKS | 9331 | 18599 | -9.166 | 0.0009 | Yes |