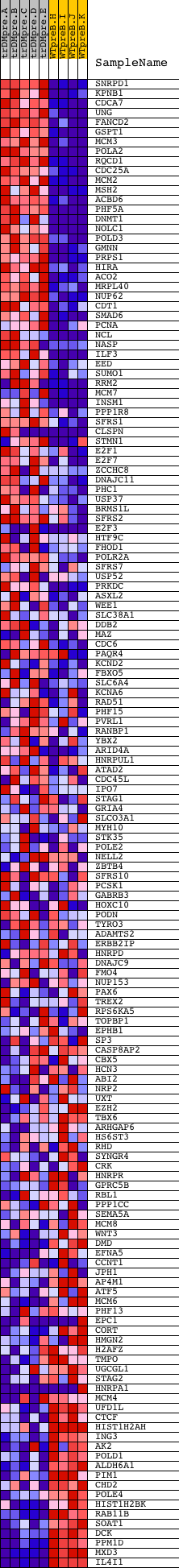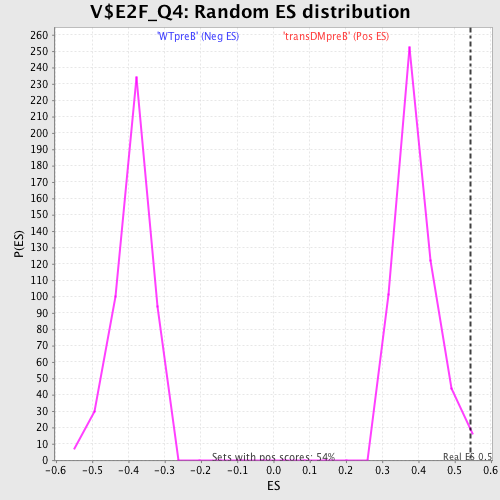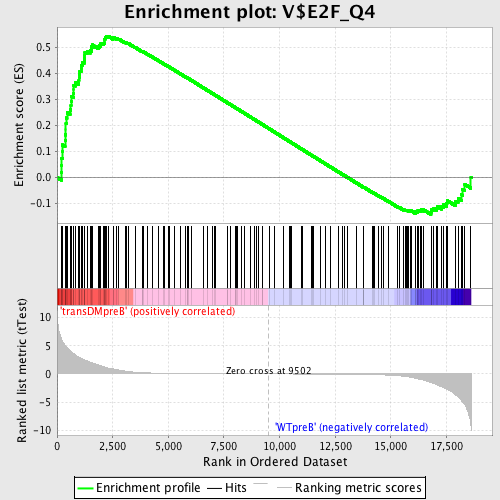
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | transDMpreB |
| GeneSet | V$E2F_Q4 |
| Enrichment Score (ES) | 0.54419726 |
| Normalized Enrichment Score (NES) | 1.3930007 |
| Nominal p-value | 0.0056074765 |
| FDR q-value | 0.24230139 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SNRPD1 | 23622 | 192 | 6.236 | 0.0182 | Yes | ||
| 2 | KPNB1 | 20274 | 202 | 6.157 | 0.0459 | Yes | ||
| 3 | CDCA7 | 12437 | 210 | 6.035 | 0.0732 | Yes | ||
| 4 | UNG | 10257 16744 | 232 | 5.883 | 0.0990 | Yes | ||
| 5 | FANCD2 | 17326 464 | 240 | 5.822 | 0.1253 | Yes | ||
| 6 | GSPT1 | 9046 | 357 | 5.076 | 0.1423 | Yes | ||
| 7 | MCM3 | 13991 | 372 | 4.986 | 0.1643 | Yes | ||
| 8 | POLA2 | 23988 | 396 | 4.885 | 0.1855 | Yes | ||
| 9 | RQCD1 | 12205 | 397 | 4.875 | 0.2078 | Yes | ||
| 10 | CDC25A | 8721 | 411 | 4.833 | 0.2293 | Yes | ||
| 11 | MCM2 | 17074 | 445 | 4.703 | 0.2490 | Yes | ||
| 12 | MSH2 | 23138 | 583 | 4.074 | 0.2603 | Yes | ||
| 13 | ACBD6 | 14089 | 621 | 3.968 | 0.2764 | Yes | ||
| 14 | PHF5A | 22194 | 627 | 3.958 | 0.2943 | Yes | ||
| 15 | DNMT1 | 19217 | 641 | 3.927 | 0.3116 | Yes | ||
| 16 | NOLC1 | 7704 | 728 | 3.609 | 0.3235 | Yes | ||
| 17 | POLD3 | 17742 | 746 | 3.578 | 0.3389 | Yes | ||
| 18 | GMNN | 21513 | 749 | 3.568 | 0.3552 | Yes | ||
| 19 | PRPS1 | 24233 | 811 | 3.405 | 0.3675 | Yes | ||
| 20 | HIRA | 4852 9090 | 979 | 2.952 | 0.3720 | Yes | ||
| 21 | ACO2 | 8527 | 984 | 2.941 | 0.3852 | Yes | ||
| 22 | MRPL40 | 22641 | 993 | 2.919 | 0.3982 | Yes | ||
| 23 | NUP62 | 9497 | 1020 | 2.854 | 0.4098 | Yes | ||
| 24 | CDT1 | 18437 | 1080 | 2.756 | 0.4192 | Yes | ||
| 25 | SMAD6 | 19083 | 1090 | 2.735 | 0.4313 | Yes | ||
| 26 | PCNA | 9535 | 1127 | 2.676 | 0.4416 | Yes | ||
| 27 | NCL | 5153 13899 | 1212 | 2.514 | 0.4486 | Yes | ||
| 28 | NASP | 2383 2387 6955 | 1225 | 2.494 | 0.4594 | Yes | ||
| 29 | ILF3 | 3110 3030 9176 | 1240 | 2.476 | 0.4699 | Yes | ||
| 30 | EED | 17759 | 1249 | 2.461 | 0.4808 | Yes | ||
| 31 | SUMO1 | 5826 3943 10247 | 1350 | 2.328 | 0.4860 | Yes | ||
| 32 | RRM2 | 5401 5400 | 1481 | 2.084 | 0.4885 | Yes | ||
| 33 | MCM7 | 9372 3568 | 1539 | 1.995 | 0.4946 | Yes | ||
| 34 | INSM1 | 14815 | 1550 | 1.979 | 0.5031 | Yes | ||
| 35 | PPP1R8 | 15730 | 1579 | 1.937 | 0.5105 | Yes | ||
| 36 | SFRS1 | 8492 | 1857 | 1.596 | 0.5028 | Yes | ||
| 37 | CLSPN | 16079 11256 6530 | 1890 | 1.549 | 0.5081 | Yes | ||
| 38 | STMN1 | 9261 | 1957 | 1.452 | 0.5112 | Yes | ||
| 39 | E2F1 | 14384 | 1966 | 1.442 | 0.5174 | Yes | ||
| 40 | E2F7 | 19884 | 2086 | 1.299 | 0.5169 | Yes | ||
| 41 | ZCCHC8 | 3570 7696 | 2131 | 1.233 | 0.5202 | Yes | ||
| 42 | DNAJC11 | 2486 6068 | 2135 | 1.231 | 0.5257 | Yes | ||
| 43 | PHC1 | 17013 | 2136 | 1.230 | 0.5313 | Yes | ||
| 44 | USP37 | 13923 | 2155 | 1.208 | 0.5358 | Yes | ||
| 45 | BRMS1L | 21268 | 2183 | 1.179 | 0.5398 | Yes | ||
| 46 | SFRS2 | 9807 20136 | 2224 | 1.131 | 0.5428 | Yes | ||
| 47 | E2F3 | 21500 8874 | 2288 | 1.048 | 0.5442 | Yes | ||
| 48 | HTF9C | 4886 1678 | 2513 | 0.889 | 0.5361 | No | ||
| 49 | FHOD1 | 18772 | 2527 | 0.882 | 0.5395 | No | ||
| 50 | POLR2A | 5394 | 2663 | 0.775 | 0.5357 | No | ||
| 51 | SFRS7 | 22889 | 2779 | 0.674 | 0.5326 | No | ||
| 52 | USP52 | 3397 19841 | 3056 | 0.489 | 0.5199 | No | ||
| 53 | PRKDC | 9617 22847 | 3126 | 0.448 | 0.5182 | No | ||
| 54 | ASXL2 | 7957 | 3193 | 0.413 | 0.5165 | No | ||
| 55 | WEE1 | 18127 | 3500 | 0.297 | 0.5013 | No | ||
| 56 | SLC38A1 | 22148 | 3837 | 0.221 | 0.4841 | No | ||
| 57 | DDB2 | 2721 2878 14525 2661 | 3844 | 0.219 | 0.4848 | No | ||
| 58 | MAZ | 1327 17623 | 3902 | 0.209 | 0.4827 | No | ||
| 59 | CDC6 | 6221 6220 | 4074 | 0.180 | 0.4742 | No | ||
| 60 | PAQR4 | 23105 | 4278 | 0.155 | 0.4639 | No | ||
| 61 | KCND2 | 4941 17515 | 4544 | 0.127 | 0.4502 | No | ||
| 62 | FBXO5 | 19830 | 4774 | 0.111 | 0.4383 | No | ||
| 63 | SLC6A4 | 20770 | 4846 | 0.107 | 0.4349 | No | ||
| 64 | KCNA6 | 664 16990 665 4938 | 5006 | 0.096 | 0.4267 | No | ||
| 65 | RAD51 | 2897 14903 | 5044 | 0.094 | 0.4252 | No | ||
| 66 | PHF15 | 20467 8050 | 5278 | 0.082 | 0.4129 | No | ||
| 67 | PVRL1 | 3006 19482 12211 7192 | 5564 | 0.069 | 0.3978 | No | ||
| 68 | RANBP1 | 9692 5357 | 5788 | 0.062 | 0.3860 | No | ||
| 69 | YBX2 | 20818 | 5844 | 0.060 | 0.3833 | No | ||
| 70 | ARID4A | 2080 6215 | 5853 | 0.060 | 0.3831 | No | ||
| 71 | HNRPUL1 | 6118 17923 1608 10577 1345 | 5894 | 0.059 | 0.3812 | No | ||
| 72 | ATAD2 | 2268 2256 | 6038 | 0.055 | 0.3738 | No | ||
| 73 | CDC45L | 22642 1752 | 6568 | 0.043 | 0.3453 | No | ||
| 74 | IPO7 | 6130 | 6750 | 0.039 | 0.3357 | No | ||
| 75 | STAG1 | 5520 9905 2987 | 7002 | 0.035 | 0.3222 | No | ||
| 76 | GRIA4 | 19249 | 7078 | 0.033 | 0.3183 | No | ||
| 77 | SLCO3A1 | 17796 4237 8430 | 7122 | 0.033 | 0.3161 | No | ||
| 78 | MYH10 | 8077 | 7677 | 0.024 | 0.2862 | No | ||
| 79 | STK35 | 14851 | 7678 | 0.024 | 0.2864 | No | ||
| 80 | POLE2 | 21053 | 7814 | 0.022 | 0.2791 | No | ||
| 81 | NELL2 | 12006 7020 22151 | 7996 | 0.020 | 0.2694 | No | ||
| 82 | ZBTB4 | 13267 | 8019 | 0.019 | 0.2683 | No | ||
| 83 | SFRS10 | 9818 1722 5441 | 8049 | 0.019 | 0.2668 | No | ||
| 84 | PCSK1 | 21600 | 8125 | 0.018 | 0.2629 | No | ||
| 85 | GABRB3 | 4747 | 8278 | 0.016 | 0.2547 | No | ||
| 86 | HOXC10 | 22342 | 8430 | 0.014 | 0.2466 | No | ||
| 87 | PODN | 15811 | 8681 | 0.010 | 0.2331 | No | ||
| 88 | TYRO3 | 5811 | 8858 | 0.008 | 0.2236 | No | ||
| 89 | ADAMTS2 | 5706 20899 1356 | 8974 | 0.007 | 0.2174 | No | ||
| 90 | ERBB2IP | 3232 12234 | 9066 | 0.006 | 0.2125 | No | ||
| 91 | HNRPD | 8645 | 9217 | 0.004 | 0.2044 | No | ||
| 92 | DNAJC9 | 21912 | 9225 | 0.004 | 0.2040 | No | ||
| 93 | FMO4 | 13786 | 9538 | -0.001 | 0.1871 | No | ||
| 94 | NUP153 | 21474 | 9748 | -0.003 | 0.1758 | No | ||
| 95 | PAX6 | 5223 | 10181 | -0.009 | 0.1525 | No | ||
| 96 | TREX2 | 24137 | 10429 | -0.012 | 0.1392 | No | ||
| 97 | RPS6KA5 | 2076 21005 | 10489 | -0.013 | 0.1360 | No | ||
| 98 | TOPBP1 | 10659 | 10535 | -0.013 | 0.1336 | No | ||
| 99 | EPHB1 | 19024 | 10967 | -0.019 | 0.1104 | No | ||
| 100 | SP3 | 5483 | 11020 | -0.020 | 0.1077 | No | ||
| 101 | CASP8AP2 | 16253 | 11427 | -0.026 | 0.0858 | No | ||
| 102 | CBX5 | 22108 4485 | 11481 | -0.027 | 0.0830 | No | ||
| 103 | HCN3 | 4843 1763 | 11507 | -0.027 | 0.0818 | No | ||
| 104 | ABI2 | 4037 6830 | 11852 | -0.033 | 0.0633 | No | ||
| 105 | NRP2 | 5194 9486 5195 | 12056 | -0.037 | 0.0525 | No | ||
| 106 | UXT | 5837 10267 | 12294 | -0.042 | 0.0399 | No | ||
| 107 | EZH2 | 1092 17163 | 12641 | -0.051 | 0.0214 | No | ||
| 108 | TBX6 | 1993 18075 | 12826 | -0.056 | 0.0116 | No | ||
| 109 | ARHGAP6 | 24207 | 12930 | -0.059 | 0.0063 | No | ||
| 110 | HS6ST3 | 21933 | 13040 | -0.063 | 0.0007 | No | ||
| 111 | RHD | 16042 | 13465 | -0.078 | -0.0219 | No | ||
| 112 | SYNGR4 | 17825 | 13769 | -0.095 | -0.0379 | No | ||
| 113 | CRK | 4559 1249 | 13780 | -0.096 | -0.0380 | No | ||
| 114 | HNRPR | 13196 7909 2500 | 13785 | -0.096 | -0.0377 | No | ||
| 115 | GPRC5B | 17662 | 14187 | -0.126 | -0.0589 | No | ||
| 116 | RBL1 | 14373 | 14231 | -0.130 | -0.0606 | No | ||
| 117 | PPP1CC | 9609 5283 | 14252 | -0.131 | -0.0611 | No | ||
| 118 | SEMA5A | 22496 5423 | 14424 | -0.149 | -0.0697 | No | ||
| 119 | MCM8 | 14834 | 14591 | -0.168 | -0.0779 | No | ||
| 120 | WNT3 | 20635 | 14670 | -0.180 | -0.0813 | No | ||
| 121 | DMD | 24295 2647 | 14880 | -0.212 | -0.0916 | No | ||
| 122 | EFNA5 | 4655 8884 | 15319 | -0.314 | -0.1139 | No | ||
| 123 | CCNT1 | 22140 11607 | 15392 | -0.339 | -0.1163 | No | ||
| 124 | JPH1 | 13994 | 15590 | -0.417 | -0.1250 | No | ||
| 125 | AP4M1 | 3499 16656 | 15643 | -0.438 | -0.1258 | No | ||
| 126 | ATF5 | 17843 | 15697 | -0.462 | -0.1266 | No | ||
| 127 | MCM6 | 4000 13845 4119 | 15750 | -0.489 | -0.1272 | No | ||
| 128 | PHF13 | 15659 | 15802 | -0.519 | -0.1276 | No | ||
| 129 | EPC1 | 2039 8907 2033 | 15863 | -0.551 | -0.1283 | No | ||
| 130 | CORT | 15669 2539 | 15920 | -0.593 | -0.1286 | No | ||
| 131 | HMGN2 | 9095 | 16119 | -0.766 | -0.1358 | No | ||
| 132 | H2AFZ | 6957 | 16123 | -0.772 | -0.1324 | No | ||
| 133 | TMPO | 5778 | 16191 | -0.856 | -0.1321 | No | ||
| 134 | UGCGL1 | 6721 | 16210 | -0.870 | -0.1291 | No | ||
| 135 | STAG2 | 5521 | 16249 | -0.911 | -0.1270 | No | ||
| 136 | HNRPA1 | 9103 4860 | 16351 | -0.999 | -0.1279 | No | ||
| 137 | MCM4 | 22655 1708 | 16371 | -1.005 | -0.1243 | No | ||
| 138 | UFD1L | 22825 | 16472 | -1.090 | -0.1248 | No | ||
| 139 | CTCF | 18490 | 16806 | -1.517 | -0.1358 | No | ||
| 140 | HIST1H2AH | 11414 | 16813 | -1.529 | -0.1292 | No | ||
| 141 | ING3 | 17514 1019 | 16839 | -1.576 | -0.1233 | No | ||
| 142 | AK2 | 8563 2479 16073 | 16931 | -1.712 | -0.1204 | No | ||
| 143 | POLD1 | 17847 | 17042 | -1.851 | -0.1178 | No | ||
| 144 | ALDH6A1 | 21025 | 17108 | -1.975 | -0.1123 | No | ||
| 145 | PIM1 | 23308 | 17289 | -2.292 | -0.1116 | No | ||
| 146 | CHD2 | 10847 | 17373 | -2.423 | -0.1050 | No | ||
| 147 | POLE4 | 12441 | 17502 | -2.639 | -0.0998 | No | ||
| 148 | HIST1H2BK | 11423 | 17527 | -2.689 | -0.0888 | No | ||
| 149 | RAB11B | 9676 | 17913 | -3.710 | -0.0926 | No | ||
| 150 | SOAT1 | 13797 | 18022 | -4.030 | -0.0800 | No | ||
| 151 | DCK | 16808 | 18181 | -4.761 | -0.0668 | No | ||
| 152 | PPM1D | 20721 | 18221 | -4.979 | -0.0461 | No | ||
| 153 | MXD3 | 21455 | 18327 | -5.489 | -0.0266 | No | ||
| 154 | IL4I1 | 8971 | 18601 | -9.209 | 0.0008 | No |