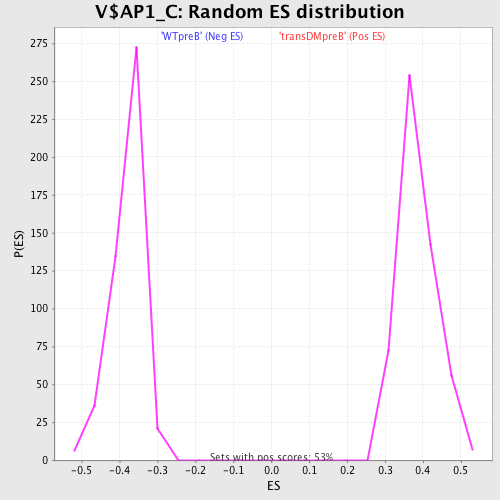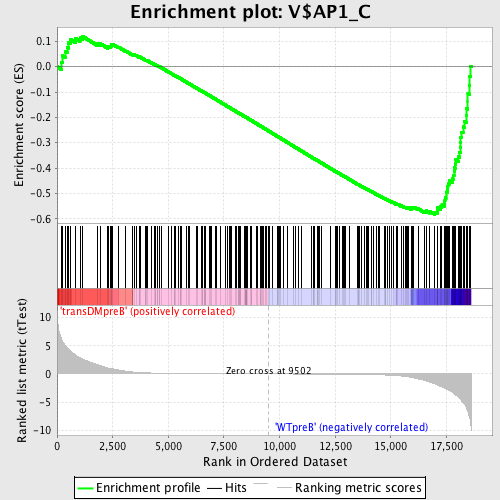
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | V$AP1_C |
| Enrichment Score (ES) | -0.58311784 |
| Normalized Enrichment Score (NES) | -1.5463686 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.08078804 |
| FWER p-Value | 0.556 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RHBDF1 | 20504 | 198 | 6.188 | 0.0180 | No | ||
| 2 | POLR3E | 18100 | 245 | 5.794 | 0.0425 | No | ||
| 3 | XPOT | 13090 | 361 | 5.055 | 0.0598 | No | ||
| 4 | SLC16A6 | 4186 | 456 | 4.614 | 0.0762 | No | ||
| 5 | FSTL1 | 22765 | 512 | 4.391 | 0.0936 | No | ||
| 6 | RBBP7 | 10887 6366 10886 | 588 | 4.070 | 0.1085 | No | ||
| 7 | SNX10 | 17445 | 828 | 3.354 | 0.1111 | No | ||
| 8 | PPP2CB | 18636 | 1030 | 2.833 | 0.1134 | No | ||
| 9 | IRAK1 | 4916 | 1128 | 2.674 | 0.1205 | No | ||
| 10 | IL10 | 14145 1510 1553 22902 | 1820 | 1.629 | 0.0906 | No | ||
| 11 | MMP9 | 14732 | 1952 | 1.461 | 0.0903 | No | ||
| 12 | LAMC1 | 13805 | 2268 | 1.069 | 0.0781 | No | ||
| 13 | SEC24D | 15429 | 2331 | 1.012 | 0.0795 | No | ||
| 14 | TYSND1 | 20007 | 2393 | 0.986 | 0.0807 | No | ||
| 15 | HDAC3 | 4844 | 2428 | 0.957 | 0.0833 | No | ||
| 16 | FBXO44 | 15675 | 2430 | 0.957 | 0.0877 | No | ||
| 17 | NFRKB | 10642 | 2480 | 0.910 | 0.0893 | No | ||
| 18 | ROCK2 | 21309 5387 | 2764 | 0.694 | 0.0772 | No | ||
| 19 | VAPA | 22908 | 3085 | 0.471 | 0.0620 | No | ||
| 20 | TLX2 | 5773 1165 | 3383 | 0.337 | 0.0474 | No | ||
| 21 | TCF7 | 1467 20466 | 3394 | 0.332 | 0.0484 | No | ||
| 22 | PKN3 | 15057 | 3409 | 0.327 | 0.0492 | No | ||
| 23 | USP2 | 19480 3052 3043 | 3489 | 0.303 | 0.0463 | No | ||
| 24 | FBXO2 | 15993 | 3564 | 0.281 | 0.0436 | No | ||
| 25 | CLSTN3 | 17006 | 3684 | 0.252 | 0.0383 | No | ||
| 26 | NEFH | 11724 | 3695 | 0.249 | 0.0389 | No | ||
| 27 | DTNA | 8870 4647 23611 1991 | 3730 | 0.242 | 0.0382 | No | ||
| 28 | MDFI | 22950 | 3979 | 0.196 | 0.0256 | No | ||
| 29 | CD68 | 4501 | 3988 | 0.195 | 0.0261 | No | ||
| 30 | TTC1 | 7342 | 4034 | 0.187 | 0.0245 | No | ||
| 31 | VASP | 5847 | 4079 | 0.179 | 0.0230 | No | ||
| 32 | FIGF | 24212 | 4248 | 0.158 | 0.0146 | No | ||
| 33 | GKN1 | 17081 | 4380 | 0.143 | 0.0081 | No | ||
| 34 | ANK3 | 8591 3445 3304 3381 | 4410 | 0.140 | 0.0072 | No | ||
| 35 | COL7A1 | 8769 4547 | 4435 | 0.138 | 0.0065 | No | ||
| 36 | EFNA1 | 15279 | 4490 | 0.133 | 0.0042 | No | ||
| 37 | RIN1 | 10349 | 4509 | 0.131 | 0.0039 | No | ||
| 38 | PDGFRB | 23539 | 4579 | 0.125 | 0.0007 | No | ||
| 39 | PCDH9 | 21736 | 4618 | 0.122 | -0.0008 | No | ||
| 40 | SNCG | 21878 | 4693 | 0.116 | -0.0043 | No | ||
| 41 | CDH23 | 3369 19759 3442 | 4988 | 0.097 | -0.0198 | No | ||
| 42 | PTPRR | 3436 19876 | 5126 | 0.089 | -0.0268 | No | ||
| 43 | IDS | 9142 2617 | 5260 | 0.083 | -0.0337 | No | ||
| 44 | RPS6KA4 | 23804 | 5296 | 0.081 | -0.0352 | No | ||
| 45 | MYOZ2 | 15174 | 5339 | 0.079 | -0.0371 | No | ||
| 46 | CMYA1 | 18966 | 5468 | 0.074 | -0.0437 | No | ||
| 47 | SHC3 | 21465 | 5545 | 0.070 | -0.0475 | No | ||
| 48 | AQP5 | 22365 | 5547 | 0.070 | -0.0473 | No | ||
| 49 | NOTCH4 | 23277 9475 | 5587 | 0.069 | -0.0491 | No | ||
| 50 | MAP1A | 5124 | 5810 | 0.061 | -0.0608 | No | ||
| 51 | FBXW11 | 20926 | 5922 | 0.058 | -0.0666 | No | ||
| 52 | EPHA1 | 17168 8908 17167 | 5937 | 0.058 | -0.0671 | No | ||
| 53 | ZFYVE9 | 10501 2449 | 6258 | 0.049 | -0.0842 | No | ||
| 54 | SMPX | 24220 | 6324 | 0.048 | -0.0875 | No | ||
| 55 | WNT7B | 22172 | 6493 | 0.044 | -0.0965 | No | ||
| 56 | CSMD3 | 10735 7938 | 6532 | 0.044 | -0.0983 | No | ||
| 57 | CSF3 | 1394 20671 | 6539 | 0.043 | -0.0985 | No | ||
| 58 | S100A5 | 15529 | 6616 | 0.042 | -0.1024 | No | ||
| 59 | TLL1 | 10188 | 6641 | 0.041 | -0.1035 | No | ||
| 60 | NTN4 | 19903 | 6650 | 0.041 | -0.1037 | No | ||
| 61 | CLDN15 | 16667 3572 | 6676 | 0.041 | -0.1049 | No | ||
| 62 | FGF11 | 20383 | 6871 | 0.037 | -0.1153 | No | ||
| 63 | EPHB2 | 4675 2440 8910 | 6902 | 0.037 | -0.1167 | No | ||
| 64 | CRYBA2 | 13920 | 6921 | 0.036 | -0.1175 | No | ||
| 65 | KBTBD5 | 13020 | 7114 | 0.033 | -0.1278 | No | ||
| 66 | MYBPH | 14130 | 7182 | 0.032 | -0.1313 | No | ||
| 67 | LTBP3 | 5008 23785 9288 | 7324 | 0.029 | -0.1388 | No | ||
| 68 | CAPN6 | 24041 | 7554 | 0.026 | -0.1512 | No | ||
| 69 | CPNE8 | 22156 | 7670 | 0.024 | -0.1573 | No | ||
| 70 | SFN | 7060 12062 | 7742 | 0.023 | -0.1611 | No | ||
| 71 | CNTF | 3741 8761 3680 3736 | 7809 | 0.022 | -0.1645 | No | ||
| 72 | ATP6V1C2 | 21098 | 7827 | 0.022 | -0.1654 | No | ||
| 73 | SH3RF2 | 23571 | 8015 | 0.019 | -0.1754 | No | ||
| 74 | TRPV3 | 20788 | 8056 | 0.019 | -0.1775 | No | ||
| 75 | ASB5 | 13322 | 8154 | 0.017 | -0.1827 | No | ||
| 76 | FLNC | 4302 8510 | 8162 | 0.017 | -0.1830 | No | ||
| 77 | SYT2 | 14122 | 8189 | 0.017 | -0.1843 | No | ||
| 78 | EYA1 | 4695 4061 | 8206 | 0.017 | -0.1851 | No | ||
| 79 | WDFY3 | 7789 3607 16462 | 8246 | 0.016 | -0.1872 | No | ||
| 80 | ABHD4 | 22021 | 8255 | 0.016 | -0.1875 | No | ||
| 81 | DIRAS1 | 9915 | 8434 | 0.014 | -0.1971 | No | ||
| 82 | SERPINB5 | 9861 | 8447 | 0.013 | -0.1977 | No | ||
| 83 | MAPRE3 | 8301 | 8459 | 0.013 | -0.1983 | No | ||
| 84 | CHST4 | 18749 | 8469 | 0.013 | -0.1987 | No | ||
| 85 | ELAVL2 | 15839 | 8501 | 0.013 | -0.2003 | No | ||
| 86 | PLCD1 | 18972 | 8510 | 0.013 | -0.2007 | No | ||
| 87 | USP13 | 13047 | 8515 | 0.013 | -0.2008 | No | ||
| 88 | RBP4 | 23686 | 8570 | 0.012 | -0.2037 | No | ||
| 89 | NUDT11 | 24387 | 8703 | 0.010 | -0.2108 | No | ||
| 90 | IL9 | 21444 | 8717 | 0.010 | -0.2115 | No | ||
| 91 | CALB2 | 8680 18748 | 8720 | 0.010 | -0.2116 | No | ||
| 92 | LOR | 15269 | 8977 | 0.007 | -0.2254 | No | ||
| 93 | XLKD1 | 17675 | 9019 | 0.006 | -0.2276 | No | ||
| 94 | FOSL1 | 23779 | 9119 | 0.005 | -0.2330 | No | ||
| 95 | PRX | 3729 9628 | 9131 | 0.005 | -0.2336 | No | ||
| 96 | WNT6 | 5881 | 9202 | 0.004 | -0.2374 | No | ||
| 97 | MNT | 20784 | 9250 | 0.003 | -0.2399 | No | ||
| 98 | LPP | 5570 | 9291 | 0.003 | -0.2421 | No | ||
| 99 | TIAL1 | 996 5753 | 9368 | 0.002 | -0.2462 | No | ||
| 100 | RB1CC1 | 4486 14299 | 9427 | 0.001 | -0.2493 | No | ||
| 101 | UBE2E3 | 5816 5815 | 9504 | -0.000 | -0.2535 | No | ||
| 102 | IBRDC2 | 5747 | 9525 | -0.000 | -0.2545 | No | ||
| 103 | OMG | 5209 | 9692 | -0.003 | -0.2635 | No | ||
| 104 | IL11 | 17981 | 9890 | -0.005 | -0.2742 | No | ||
| 105 | PHLDA2 | 17545 | 9898 | -0.005 | -0.2746 | No | ||
| 106 | MCF2 | 24147 | 9951 | -0.006 | -0.2774 | No | ||
| 107 | GDNF | 22514 2275 | 9982 | -0.006 | -0.2790 | No | ||
| 108 | STAT5B | 20222 | 10037 | -0.007 | -0.2819 | No | ||
| 109 | SFTPC | 21759 | 10160 | -0.009 | -0.2885 | No | ||
| 110 | PROCR | 14772 | 10342 | -0.011 | -0.2982 | No | ||
| 111 | HEPH | 24288 2623 2635 2568 2652 | 10640 | -0.015 | -0.3143 | No | ||
| 112 | SCEL | 21942 | 10696 | -0.015 | -0.3172 | No | ||
| 113 | VIT | 23155 | 10851 | -0.018 | -0.3255 | No | ||
| 114 | SLCO5A1 | 6286 | 10974 | -0.019 | -0.3320 | No | ||
| 115 | CAPN12 | 18314 | 11437 | -0.026 | -0.3570 | No | ||
| 116 | HCN3 | 4843 1763 | 11507 | -0.027 | -0.3606 | No | ||
| 117 | MPP5 | 21230 7078 | 11542 | -0.028 | -0.3624 | No | ||
| 118 | MARK4 | 10572 | 11577 | -0.029 | -0.3641 | No | ||
| 119 | ADAM15 | 15275 | 11700 | -0.031 | -0.3706 | No | ||
| 120 | LAMC2 | 4983 9266 13806 3975 | 11703 | -0.031 | -0.3705 | No | ||
| 121 | PRDM1 | 19775 3337 | 11762 | -0.032 | -0.3735 | No | ||
| 122 | APBA2BP | 14385 | 11779 | -0.032 | -0.3742 | No | ||
| 123 | MAMDC2 | 23711 | 11885 | -0.034 | -0.3798 | No | ||
| 124 | ESRRG | 6447 | 12274 | -0.042 | -0.4007 | No | ||
| 125 | MARK1 | 5955 | 12499 | -0.048 | -0.4126 | No | ||
| 126 | IVL | 15263 | 12531 | -0.049 | -0.4141 | No | ||
| 127 | HOXA11 | 17146 | 12543 | -0.049 | -0.4145 | No | ||
| 128 | PLEC1 | 2273 2198 2205 2178 2230 22247 2213 2231 2295 2172 5263 2266 | 12586 | -0.050 | -0.4165 | No | ||
| 129 | PACSIN3 | 14948 8149 2776 2665 | 12704 | -0.053 | -0.4226 | No | ||
| 130 | LMNA | 4999 15290 1778 9280 | 12847 | -0.056 | -0.4301 | No | ||
| 131 | ANKRD22 | 6970 | 12853 | -0.056 | -0.4301 | No | ||
| 132 | SNCB | 21456 | 12925 | -0.059 | -0.4337 | No | ||
| 133 | SLC38A3 | 13319 | 12965 | -0.060 | -0.4355 | No | ||
| 134 | SLC26A1 | 16446 | 13129 | -0.066 | -0.4441 | No | ||
| 135 | VDR | 5852 10285 | 13506 | -0.081 | -0.4641 | No | ||
| 136 | DSCR1L1 | 23231 | 13539 | -0.082 | -0.4655 | No | ||
| 137 | SEMA3B | 5422 | 13563 | -0.083 | -0.4663 | No | ||
| 138 | MET | 17520 | 13610 | -0.086 | -0.4684 | No | ||
| 139 | SYNGR1 | 22420 2290 | 13672 | -0.089 | -0.4713 | No | ||
| 140 | HSPB7 | 16005 | 13833 | -0.100 | -0.4795 | No | ||
| 141 | FABP4 | 8601 | 13924 | -0.107 | -0.4839 | No | ||
| 142 | SLITRK6 | 21724 | 13944 | -0.108 | -0.4845 | No | ||
| 143 | COL16A1 | 16066 | 13977 | -0.110 | -0.4857 | No | ||
| 144 | HSPB8 | 16405 | 14116 | -0.119 | -0.4926 | No | ||
| 145 | DYRK1A | 4649 | 14123 | -0.120 | -0.4924 | No | ||
| 146 | CLDN17 | 10757 | 14213 | -0.128 | -0.4966 | No | ||
| 147 | TUBB4 | 22917 | 14355 | -0.141 | -0.5037 | No | ||
| 148 | LAMA3 | 23620 2009 | 14454 | -0.152 | -0.5083 | No | ||
| 149 | SLC22A18 | 18000 | 14506 | -0.157 | -0.5103 | No | ||
| 150 | AIG1 | 7272 | 14722 | -0.187 | -0.5211 | No | ||
| 151 | PITPNC1 | 7759 | 14752 | -0.191 | -0.5218 | No | ||
| 152 | LENEP | 12181 | 14756 | -0.191 | -0.5211 | No | ||
| 153 | ADRA1A | 2850 8560 | 14847 | -0.205 | -0.5250 | No | ||
| 154 | SPRED1 | 4331 | 14947 | -0.225 | -0.5293 | No | ||
| 155 | FGF9 | 8966 | 15041 | -0.246 | -0.5333 | No | ||
| 156 | RICS | 19523 | 15111 | -0.260 | -0.5358 | No | ||
| 157 | MYB | 3344 3355 19806 | 15237 | -0.290 | -0.5412 | No | ||
| 158 | BAZ2A | 4371 | 15264 | -0.299 | -0.5413 | No | ||
| 159 | XYLT1 | 18113 | 15314 | -0.313 | -0.5425 | No | ||
| 160 | SV2B | 17795 | 15464 | -0.365 | -0.5489 | No | ||
| 161 | BLMH | 20769 | 15554 | -0.405 | -0.5518 | No | ||
| 162 | ULK1 | 16436 | 15641 | -0.437 | -0.5544 | No | ||
| 163 | SYTL1 | 15728 | 15716 | -0.471 | -0.5563 | No | ||
| 164 | GRIA1 | 20877 4801 9040 | 15743 | -0.485 | -0.5554 | No | ||
| 165 | VAT1 | 11253 | 15777 | -0.499 | -0.5549 | No | ||
| 166 | VCL | 22083 | 15915 | -0.589 | -0.5596 | No | ||
| 167 | ADCK4 | 13364 | 15922 | -0.595 | -0.5572 | No | ||
| 168 | ROM1 | 23751 | 15969 | -0.632 | -0.5567 | No | ||
| 169 | TRAPPC3 | 6553 16080 2469 11305 | 15990 | -0.653 | -0.5548 | No | ||
| 170 | ABCF3 | 6578 | 16032 | -0.685 | -0.5538 | No | ||
| 171 | BTK | 24061 | 16235 | -0.895 | -0.5606 | No | ||
| 172 | DTX2 | 16674 3488 | 16497 | -1.120 | -0.5696 | No | ||
| 173 | LRRFIP2 | 3024 3071 19285 | 16585 | -1.208 | -0.5687 | No | ||
| 174 | CENTD1 | 16529 | 16746 | -1.439 | -0.5707 | No | ||
| 175 | SDCBP | 2339 6998 6999 | 16976 | -1.750 | -0.5750 | Yes | ||
| 176 | MCART1 | 10492 | 17083 | -1.929 | -0.5718 | Yes | ||
| 177 | FBS1 | 18067 | 17092 | -1.944 | -0.5631 | Yes | ||
| 178 | AP2M1 | 8603 | 17114 | -1.980 | -0.5551 | Yes | ||
| 179 | SSH2 | 6202 | 17246 | -2.211 | -0.5519 | Yes | ||
| 180 | PPP2CA | 20890 | 17299 | -2.304 | -0.5440 | Yes | ||
| 181 | PEA15 | 13753 | 17410 | -2.499 | -0.5383 | Yes | ||
| 182 | CLIC1 | 23262 | 17429 | -2.526 | -0.5276 | Yes | ||
| 183 | TOB1 | 20703 | 17438 | -2.536 | -0.5162 | Yes | ||
| 184 | BTG3 | 8664 | 17482 | -2.592 | -0.5065 | Yes | ||
| 185 | GADD45G | 21637 | 17483 | -2.597 | -0.4944 | Yes | ||
| 186 | BACH1 | 8647 | 17544 | -2.722 | -0.4850 | Yes | ||
| 187 | EPHA2 | 16006 | 17560 | -2.758 | -0.4730 | Yes | ||
| 188 | TBC1D17 | 6121 | 17571 | -2.780 | -0.4606 | Yes | ||
| 189 | GABARAPL1 | 17268 | 17640 | -2.923 | -0.4506 | Yes | ||
| 190 | RAB3D | 19205 | 17763 | -3.231 | -0.4422 | Yes | ||
| 191 | RELA | 23783 | 17808 | -3.389 | -0.4289 | Yes | ||
| 192 | USP3 | 19074 | 17844 | -3.511 | -0.4144 | Yes | ||
| 193 | BLR1 | 19145 3154 | 17862 | -3.555 | -0.3988 | Yes | ||
| 194 | ATP6V1B2 | 18599 | 17891 | -3.657 | -0.3833 | Yes | ||
| 195 | CMAS | 17245 | 17923 | -3.740 | -0.3676 | Yes | ||
| 196 | ETV5 | 22630 | 18044 | -4.102 | -0.3550 | Yes | ||
| 197 | RGS2 | 5379 | 18079 | -4.250 | -0.3371 | Yes | ||
| 198 | ITPKC | 17919 | 18117 | -4.408 | -0.3186 | Yes | ||
| 199 | PSME4 | 8339 20934 | 18128 | -4.473 | -0.2983 | Yes | ||
| 200 | PPP1R9B | 20698 | 18139 | -4.519 | -0.2778 | Yes | ||
| 201 | RNF34 | 13479 16710 3637 | 18180 | -4.745 | -0.2579 | Yes | ||
| 202 | EMP1 | 17260 | 18249 | -5.131 | -0.2377 | Yes | ||
| 203 | GADD45A | 17129 | 18325 | -5.480 | -0.2163 | Yes | ||
| 204 | RIT1 | 5382 | 18388 | -5.891 | -0.1922 | Yes | ||
| 205 | GJA1 | 20023 | 18396 | -5.981 | -0.1648 | Yes | ||
| 206 | ABCD1 | 24305 | 18464 | -6.698 | -0.1372 | Yes | ||
| 207 | POLD4 | 12822 | 18466 | -6.742 | -0.1059 | Yes | ||
| 208 | ATP1B1 | 4420 | 18542 | -7.769 | -0.0738 | Yes | ||
| 209 | LAPTM5 | 16061 | 18553 | -7.893 | -0.0376 | Yes | ||
| 210 | SLC35B1 | 1631 23226 | 18586 | -8.808 | 0.0016 | Yes |