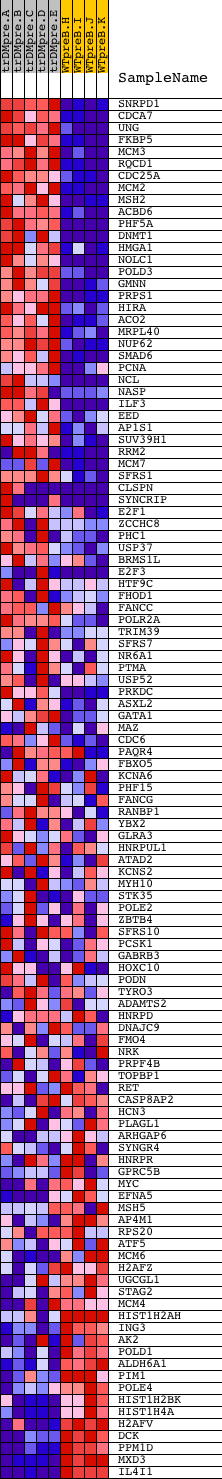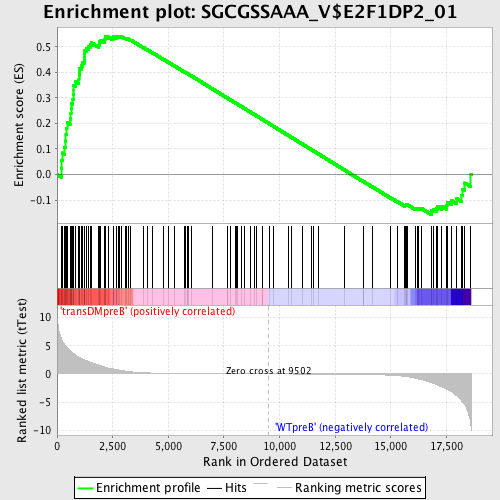
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | transDMpreB |
| GeneSet | SGCGSSAAA_V$E2F1DP2_01 |
| Enrichment Score (ES) | 0.5415655 |
| Normalized Enrichment Score (NES) | 1.3484205 |
| Nominal p-value | 0.02443609 |
| FDR q-value | 0.24227951 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SNRPD1 | 23622 | 192 | 6.236 | 0.0228 | Yes | ||
| 2 | CDCA7 | 12437 | 210 | 6.035 | 0.0540 | Yes | ||
| 3 | UNG | 10257 16744 | 232 | 5.883 | 0.0841 | Yes | ||
| 4 | FKBP5 | 23051 | 329 | 5.237 | 0.1068 | Yes | ||
| 5 | MCM3 | 13991 | 372 | 4.986 | 0.1310 | Yes | ||
| 6 | RQCD1 | 12205 | 397 | 4.875 | 0.1556 | Yes | ||
| 7 | CDC25A | 8721 | 411 | 4.833 | 0.1806 | Yes | ||
| 8 | MCM2 | 17074 | 445 | 4.703 | 0.2039 | Yes | ||
| 9 | MSH2 | 23138 | 583 | 4.074 | 0.2181 | Yes | ||
| 10 | ACBD6 | 14089 | 621 | 3.968 | 0.2372 | Yes | ||
| 11 | PHF5A | 22194 | 627 | 3.958 | 0.2580 | Yes | ||
| 12 | DNMT1 | 19217 | 641 | 3.927 | 0.2782 | Yes | ||
| 13 | HMGA1 | 23323 | 710 | 3.664 | 0.2940 | Yes | ||
| 14 | NOLC1 | 7704 | 728 | 3.609 | 0.3123 | Yes | ||
| 15 | POLD3 | 17742 | 746 | 3.578 | 0.3304 | Yes | ||
| 16 | GMNN | 21513 | 749 | 3.568 | 0.3492 | Yes | ||
| 17 | PRPS1 | 24233 | 811 | 3.405 | 0.3641 | Yes | ||
| 18 | HIRA | 4852 9090 | 979 | 2.952 | 0.3707 | Yes | ||
| 19 | ACO2 | 8527 | 984 | 2.941 | 0.3861 | Yes | ||
| 20 | MRPL40 | 22641 | 993 | 2.919 | 0.4012 | Yes | ||
| 21 | NUP62 | 9497 | 1020 | 2.854 | 0.4150 | Yes | ||
| 22 | SMAD6 | 19083 | 1090 | 2.735 | 0.4258 | Yes | ||
| 23 | PCNA | 9535 | 1127 | 2.676 | 0.4381 | Yes | ||
| 24 | NCL | 5153 13899 | 1212 | 2.514 | 0.4469 | Yes | ||
| 25 | NASP | 2383 2387 6955 | 1225 | 2.494 | 0.4596 | Yes | ||
| 26 | ILF3 | 3110 3030 9176 | 1240 | 2.476 | 0.4720 | Yes | ||
| 27 | EED | 17759 | 1249 | 2.461 | 0.4846 | Yes | ||
| 28 | AP1S1 | 3500 3453 16335 | 1306 | 2.379 | 0.4942 | Yes | ||
| 29 | SUV39H1 | 24194 | 1410 | 2.217 | 0.5005 | Yes | ||
| 30 | RRM2 | 5401 5400 | 1481 | 2.084 | 0.5078 | Yes | ||
| 31 | MCM7 | 9372 3568 | 1539 | 1.995 | 0.5153 | Yes | ||
| 32 | SFRS1 | 8492 | 1857 | 1.596 | 0.5066 | Yes | ||
| 33 | CLSPN | 16079 11256 6530 | 1890 | 1.549 | 0.5132 | Yes | ||
| 34 | SYNCRIP | 3078 3035 3107 | 1899 | 1.539 | 0.5209 | Yes | ||
| 35 | E2F1 | 14384 | 1966 | 1.442 | 0.5250 | Yes | ||
| 36 | ZCCHC8 | 3570 7696 | 2131 | 1.233 | 0.5227 | Yes | ||
| 37 | PHC1 | 17013 | 2136 | 1.230 | 0.5290 | Yes | ||
| 38 | USP37 | 13923 | 2155 | 1.208 | 0.5345 | Yes | ||
| 39 | BRMS1L | 21268 | 2183 | 1.179 | 0.5393 | Yes | ||
| 40 | E2F3 | 21500 8874 | 2288 | 1.048 | 0.5392 | Yes | ||
| 41 | HTF9C | 4886 1678 | 2513 | 0.889 | 0.5319 | Yes | ||
| 42 | FHOD1 | 18772 | 2527 | 0.882 | 0.5359 | Yes | ||
| 43 | FANCC | 4712 | 2538 | 0.873 | 0.5400 | Yes | ||
| 44 | POLR2A | 5394 | 2663 | 0.775 | 0.5374 | Yes | ||
| 45 | TRIM39 | 22994 | 2667 | 0.772 | 0.5413 | Yes | ||
| 46 | SFRS7 | 22889 | 2779 | 0.674 | 0.5389 | Yes | ||
| 47 | NR6A1 | 14596 | 2796 | 0.662 | 0.5416 | Yes | ||
| 48 | PTMA | 9657 | 2914 | 0.576 | 0.5383 | No | ||
| 49 | USP52 | 3397 19841 | 3056 | 0.489 | 0.5333 | No | ||
| 50 | PRKDC | 9617 22847 | 3126 | 0.448 | 0.5319 | No | ||
| 51 | ASXL2 | 7957 | 3193 | 0.413 | 0.5306 | No | ||
| 52 | GATA1 | 24196 | 3313 | 0.366 | 0.5261 | No | ||
| 53 | MAZ | 1327 17623 | 3902 | 0.209 | 0.4954 | No | ||
| 54 | CDC6 | 6221 6220 | 4074 | 0.180 | 0.4871 | No | ||
| 55 | PAQR4 | 23105 | 4278 | 0.155 | 0.4770 | No | ||
| 56 | FBXO5 | 19830 | 4774 | 0.111 | 0.4508 | No | ||
| 57 | KCNA6 | 664 16990 665 4938 | 5006 | 0.096 | 0.4388 | No | ||
| 58 | PHF15 | 20467 8050 | 5278 | 0.082 | 0.4246 | No | ||
| 59 | FANCG | 15904 | 5732 | 0.063 | 0.4005 | No | ||
| 60 | RANBP1 | 9692 5357 | 5788 | 0.062 | 0.3978 | No | ||
| 61 | YBX2 | 20818 | 5844 | 0.060 | 0.3952 | No | ||
| 62 | GLRA3 | 18617 | 5865 | 0.060 | 0.3944 | No | ||
| 63 | HNRPUL1 | 6118 17923 1608 10577 1345 | 5894 | 0.059 | 0.3932 | No | ||
| 64 | ATAD2 | 2268 2256 | 6038 | 0.055 | 0.3858 | No | ||
| 65 | KCNS2 | 4949 2232 | 7006 | 0.035 | 0.3337 | No | ||
| 66 | MYH10 | 8077 | 7677 | 0.024 | 0.2976 | No | ||
| 67 | STK35 | 14851 | 7678 | 0.024 | 0.2977 | No | ||
| 68 | POLE2 | 21053 | 7814 | 0.022 | 0.2906 | No | ||
| 69 | ZBTB4 | 13267 | 8019 | 0.019 | 0.2796 | No | ||
| 70 | SFRS10 | 9818 1722 5441 | 8049 | 0.019 | 0.2782 | No | ||
| 71 | PCSK1 | 21600 | 8125 | 0.018 | 0.2742 | No | ||
| 72 | GABRB3 | 4747 | 8278 | 0.016 | 0.2661 | No | ||
| 73 | HOXC10 | 22342 | 8430 | 0.014 | 0.2580 | No | ||
| 74 | PODN | 15811 | 8681 | 0.010 | 0.2445 | No | ||
| 75 | TYRO3 | 5811 | 8858 | 0.008 | 0.2351 | No | ||
| 76 | ADAMTS2 | 5706 20899 1356 | 8974 | 0.007 | 0.2289 | No | ||
| 77 | HNRPD | 8645 | 9217 | 0.004 | 0.2158 | No | ||
| 78 | DNAJC9 | 21912 | 9225 | 0.004 | 0.2155 | No | ||
| 79 | FMO4 | 13786 | 9538 | -0.001 | 0.1986 | No | ||
| 80 | NRK | 6559 | 9714 | -0.003 | 0.1892 | No | ||
| 81 | PRPF4B | 3208 5295 | 10398 | -0.012 | 0.1523 | No | ||
| 82 | TOPBP1 | 10659 | 10535 | -0.013 | 0.1450 | No | ||
| 83 | RET | 17028 | 11047 | -0.021 | 0.1175 | No | ||
| 84 | CASP8AP2 | 16253 | 11427 | -0.026 | 0.0972 | No | ||
| 85 | HCN3 | 4843 1763 | 11507 | -0.027 | 0.0931 | No | ||
| 86 | PLAGL1 | 10372 3414 | 11764 | -0.032 | 0.0794 | No | ||
| 87 | ARHGAP6 | 24207 | 12930 | -0.059 | 0.0167 | No | ||
| 88 | SYNGR4 | 17825 | 13769 | -0.095 | -0.0280 | No | ||
| 89 | HNRPR | 13196 7909 2500 | 13785 | -0.096 | -0.0283 | No | ||
| 90 | GPRC5B | 17662 | 14187 | -0.126 | -0.0494 | No | ||
| 91 | MYC | 22465 9435 | 14983 | -0.233 | -0.0911 | No | ||
| 92 | EFNA5 | 4655 8884 | 15319 | -0.314 | -0.1075 | No | ||
| 93 | MSH5 | 23010 1599 | 15631 | -0.435 | -0.1220 | No | ||
| 94 | AP4M1 | 3499 16656 | 15643 | -0.438 | -0.1203 | No | ||
| 95 | RPS20 | 7438 | 15692 | -0.460 | -0.1204 | No | ||
| 96 | ATF5 | 17843 | 15697 | -0.462 | -0.1182 | No | ||
| 97 | MCM6 | 4000 13845 4119 | 15750 | -0.489 | -0.1184 | No | ||
| 98 | H2AFZ | 6957 | 16123 | -0.772 | -0.1344 | No | ||
| 99 | UGCGL1 | 6721 | 16210 | -0.870 | -0.1344 | No | ||
| 100 | STAG2 | 5521 | 16249 | -0.911 | -0.1316 | No | ||
| 101 | MCM4 | 22655 1708 | 16371 | -1.005 | -0.1328 | No | ||
| 102 | HIST1H2AH | 11414 | 16813 | -1.529 | -0.1485 | No | ||
| 103 | ING3 | 17514 1019 | 16839 | -1.576 | -0.1415 | No | ||
| 104 | AK2 | 8563 2479 16073 | 16931 | -1.712 | -0.1373 | No | ||
| 105 | POLD1 | 17847 | 17042 | -1.851 | -0.1334 | No | ||
| 106 | ALDH6A1 | 21025 | 17108 | -1.975 | -0.1264 | No | ||
| 107 | PIM1 | 23308 | 17289 | -2.292 | -0.1240 | No | ||
| 108 | POLE4 | 12441 | 17502 | -2.639 | -0.1214 | No | ||
| 109 | HIST1H2BK | 11423 | 17527 | -2.689 | -0.1084 | No | ||
| 110 | HIST1H4A | 11590 | 17734 | -3.148 | -0.1028 | No | ||
| 111 | H2AFV | 1273 20531 | 17949 | -3.802 | -0.0941 | No | ||
| 112 | DCK | 16808 | 18181 | -4.761 | -0.0813 | No | ||
| 113 | PPM1D | 20721 | 18221 | -4.979 | -0.0569 | No | ||
| 114 | MXD3 | 21455 | 18327 | -5.489 | -0.0334 | No | ||
| 115 | IL4I1 | 8971 | 18601 | -9.209 | 0.0008 | No |