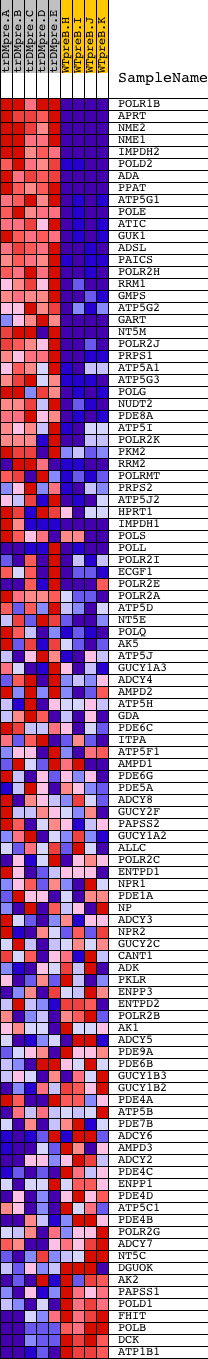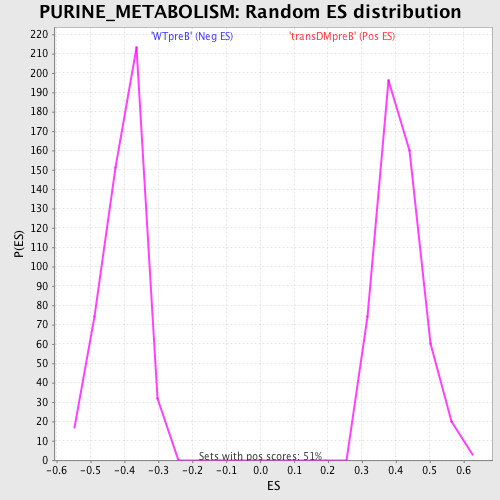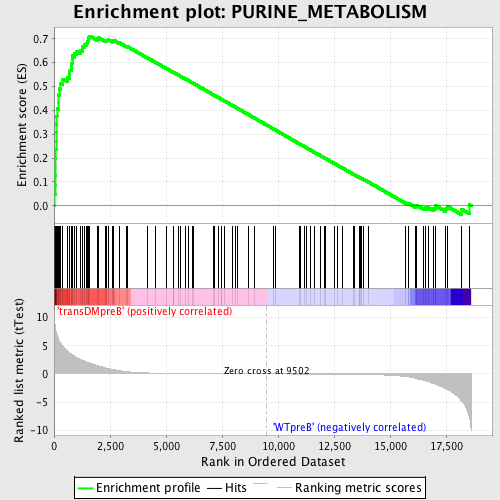
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | transDMpreB |
| GeneSet | PURINE_METABOLISM |
| Enrichment Score (ES) | 0.7112742 |
| Normalized Enrichment Score (NES) | 1.7302337 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.014840487 |
| FWER p-Value | 0.02 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | POLR1B | 14857 | 9 | 10.065 | 0.0475 | Yes | ||
| 2 | APRT | 8620 | 44 | 8.604 | 0.0867 | Yes | ||
| 3 | NME2 | 9468 | 49 | 8.321 | 0.1261 | Yes | ||
| 4 | NME1 | 9467 | 68 | 7.855 | 0.1626 | Yes | ||
| 5 | IMPDH2 | 10730 | 69 | 7.844 | 0.2000 | Yes | ||
| 6 | POLD2 | 20537 | 78 | 7.693 | 0.2362 | Yes | ||
| 7 | ADA | 2703 14361 | 89 | 7.521 | 0.2715 | Yes | ||
| 8 | PPAT | 6081 | 93 | 7.493 | 0.3071 | Yes | ||
| 9 | ATP5G1 | 8636 | 101 | 7.285 | 0.3414 | Yes | ||
| 10 | POLE | 16755 | 111 | 7.095 | 0.3748 | Yes | ||
| 11 | ATIC | 14231 3968 | 130 | 6.917 | 0.4068 | Yes | ||
| 12 | GUK1 | 20432 | 179 | 6.377 | 0.4346 | Yes | ||
| 13 | ADSL | 4358 | 181 | 6.342 | 0.4648 | Yes | ||
| 14 | PAICS | 16820 | 226 | 5.909 | 0.4906 | Yes | ||
| 15 | POLR2H | 10888 | 296 | 5.463 | 0.5129 | Yes | ||
| 16 | RRM1 | 18163 | 391 | 4.897 | 0.5311 | Yes | ||
| 17 | GMPS | 15578 | 598 | 4.048 | 0.5393 | Yes | ||
| 18 | ATP5G2 | 12610 | 681 | 3.781 | 0.5529 | Yes | ||
| 19 | GART | 22543 1754 | 706 | 3.684 | 0.5692 | Yes | ||
| 20 | NT5M | 8345 4175 | 778 | 3.508 | 0.5820 | Yes | ||
| 21 | POLR2J | 16672 | 793 | 3.452 | 0.5977 | Yes | ||
| 22 | PRPS1 | 24233 | 811 | 3.405 | 0.6131 | Yes | ||
| 23 | ATP5A1 | 23505 | 815 | 3.395 | 0.6291 | Yes | ||
| 24 | ATP5G3 | 14558 | 918 | 3.114 | 0.6384 | Yes | ||
| 25 | POLG | 17789 | 1011 | 2.873 | 0.6471 | Yes | ||
| 26 | NUDT2 | 16239 | 1174 | 2.584 | 0.6507 | Yes | ||
| 27 | PDE8A | 18202 | 1273 | 2.426 | 0.6570 | Yes | ||
| 28 | ATP5I | 8637 | 1283 | 2.411 | 0.6680 | Yes | ||
| 29 | POLR2K | 9413 | 1342 | 2.339 | 0.6760 | Yes | ||
| 30 | PKM2 | 3642 9573 | 1435 | 2.171 | 0.6814 | Yes | ||
| 31 | RRM2 | 5401 5400 | 1481 | 2.084 | 0.6889 | Yes | ||
| 32 | POLRMT | 19705 | 1515 | 2.036 | 0.6968 | Yes | ||
| 33 | PRPS2 | 24003 | 1523 | 2.022 | 0.7061 | Yes | ||
| 34 | ATP5J2 | 12186 | 1596 | 1.907 | 0.7113 | Yes | ||
| 35 | HPRT1 | 2655 24339 408 | 1927 | 1.506 | 0.7006 | No | ||
| 36 | IMPDH1 | 17197 1131 | 1971 | 1.436 | 0.7052 | No | ||
| 37 | POLS | 9963 | 2281 | 1.051 | 0.6935 | No | ||
| 38 | POLL | 23658 3688 | 2349 | 1.002 | 0.6946 | No | ||
| 39 | POLR2I | 12839 | 2417 | 0.967 | 0.6956 | No | ||
| 40 | ECGF1 | 22160 | 2592 | 0.833 | 0.6902 | No | ||
| 41 | POLR2E | 3325 19699 | 2644 | 0.791 | 0.6912 | No | ||
| 42 | POLR2A | 5394 | 2663 | 0.775 | 0.6939 | No | ||
| 43 | ATP5D | 19949 | 2903 | 0.583 | 0.6838 | No | ||
| 44 | NT5E | 19360 18702 | 3241 | 0.392 | 0.6675 | No | ||
| 45 | POLQ | 13407 22768 | 3279 | 0.380 | 0.6673 | No | ||
| 46 | AK5 | 6042 | 4178 | 0.166 | 0.6196 | No | ||
| 47 | ATP5J | 880 22555 | 4543 | 0.127 | 0.6005 | No | ||
| 48 | GUCY1A3 | 7216 12245 15310 | 5030 | 0.095 | 0.5747 | No | ||
| 49 | ADCY4 | 2980 21818 | 5320 | 0.079 | 0.5595 | No | ||
| 50 | AMPD2 | 1931 15197 | 5322 | 0.079 | 0.5598 | No | ||
| 51 | ATP5H | 12948 | 5552 | 0.070 | 0.5478 | No | ||
| 52 | GDA | 4764 | 5663 | 0.066 | 0.5421 | No | ||
| 53 | PDE6C | 8496 23868 | 5883 | 0.059 | 0.5306 | No | ||
| 54 | ITPA | 9193 | 5988 | 0.056 | 0.5252 | No | ||
| 55 | ATP5F1 | 15212 | 6180 | 0.052 | 0.5152 | No | ||
| 56 | AMPD1 | 5051 | 6243 | 0.050 | 0.5120 | No | ||
| 57 | PDE6G | 20119 | 7118 | 0.033 | 0.4650 | No | ||
| 58 | PDE5A | 15431 | 7149 | 0.032 | 0.4635 | No | ||
| 59 | ADCY8 | 22281 | 7337 | 0.029 | 0.4536 | No | ||
| 60 | GUCY2F | 24045 | 7454 | 0.027 | 0.4474 | No | ||
| 61 | PAPSS2 | 6258 | 7461 | 0.027 | 0.4472 | No | ||
| 62 | GUCY1A2 | 19580 | 7613 | 0.025 | 0.4392 | No | ||
| 63 | ALLC | 21093 2130 | 7947 | 0.020 | 0.4213 | No | ||
| 64 | POLR2C | 9750 | 8109 | 0.018 | 0.4127 | No | ||
| 65 | ENTPD1 | 4495 | 8186 | 0.017 | 0.4087 | No | ||
| 66 | NPR1 | 9480 | 8679 | 0.010 | 0.3821 | No | ||
| 67 | PDE1A | 9541 5234 | 8950 | 0.007 | 0.3676 | No | ||
| 68 | NP | 22027 9597 5273 5274 | 9774 | -0.004 | 0.3231 | No | ||
| 69 | ADCY3 | 21329 894 | 9900 | -0.005 | 0.3164 | No | ||
| 70 | NPR2 | 16230 | 10933 | -0.019 | 0.2608 | No | ||
| 71 | GUCY2C | 1140 16954 | 11013 | -0.020 | 0.2566 | No | ||
| 72 | CANT1 | 13304 | 11171 | -0.022 | 0.2482 | No | ||
| 73 | ADK | 3302 3454 8555 | 11262 | -0.023 | 0.2435 | No | ||
| 74 | PKLR | 1850 15545 | 11428 | -0.026 | 0.2347 | No | ||
| 75 | ENPP3 | 19803 | 11642 | -0.030 | 0.2233 | No | ||
| 76 | ENTPD2 | 8713 | 11875 | -0.034 | 0.2109 | No | ||
| 77 | POLR2B | 16817 | 12062 | -0.037 | 0.2011 | No | ||
| 78 | AK1 | 4363 | 12129 | -0.039 | 0.1977 | No | ||
| 79 | ADCY5 | 10312 | 12495 | -0.047 | 0.1782 | No | ||
| 80 | PDE9A | 23301 | 12656 | -0.052 | 0.1698 | No | ||
| 81 | PDE6B | 16760 | 12886 | -0.057 | 0.1577 | No | ||
| 82 | GUCY1B3 | 15311 | 13357 | -0.074 | 0.1327 | No | ||
| 83 | GUCY1B2 | 21794 | 13410 | -0.076 | 0.1302 | No | ||
| 84 | PDE4A | 3037 19542 3083 3023 | 13623 | -0.087 | 0.1192 | No | ||
| 85 | ATP5B | 19846 | 13631 | -0.088 | 0.1192 | No | ||
| 86 | PDE7B | 19807 | 13696 | -0.091 | 0.1162 | No | ||
| 87 | ADCY6 | 22139 2283 8551 | 13737 | -0.094 | 0.1145 | No | ||
| 88 | AMPD3 | 4383 2110 | 13809 | -0.098 | 0.1111 | No | ||
| 89 | ADCY2 | 21412 | 14026 | -0.113 | 0.1000 | No | ||
| 90 | PDE4C | 8481 | 15676 | -0.453 | 0.0131 | No | ||
| 91 | ENPP1 | 19804 | 15800 | -0.517 | 0.0089 | No | ||
| 92 | PDE4D | 10722 6235 | 15809 | -0.523 | 0.0110 | No | ||
| 93 | ATP5C1 | 8635 | 16131 | -0.780 | -0.0027 | No | ||
| 94 | PDE4B | 9543 | 16149 | -0.808 | 0.0003 | No | ||
| 95 | POLR2G | 23753 | 16179 | -0.839 | 0.0027 | No | ||
| 96 | ADCY7 | 8552 448 | 16511 | -1.133 | -0.0098 | No | ||
| 97 | NT5C | 20151 | 16582 | -1.203 | -0.0078 | No | ||
| 98 | DGUOK | 17099 1041 | 16691 | -1.355 | -0.0072 | No | ||
| 99 | AK2 | 8563 2479 16073 | 16931 | -1.712 | -0.0120 | No | ||
| 100 | PAPSS1 | 15419 | 17023 | -1.824 | -0.0082 | No | ||
| 101 | POLD1 | 17847 | 17042 | -1.851 | -0.0003 | No | ||
| 102 | FHIT | 21920 | 17491 | -2.617 | -0.0120 | No | ||
| 103 | POLB | 9599 | 17541 | -2.711 | -0.0018 | No | ||
| 104 | DCK | 16808 | 18181 | -4.761 | -0.0136 | No | ||
| 105 | ATP1B1 | 4420 | 18542 | -7.769 | 0.0040 | No |