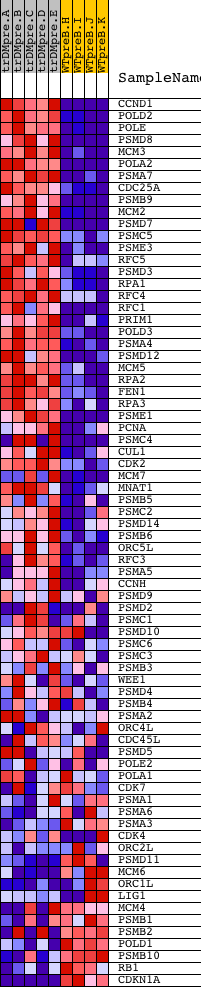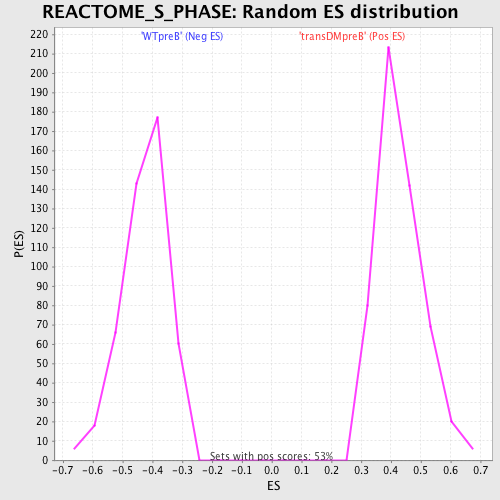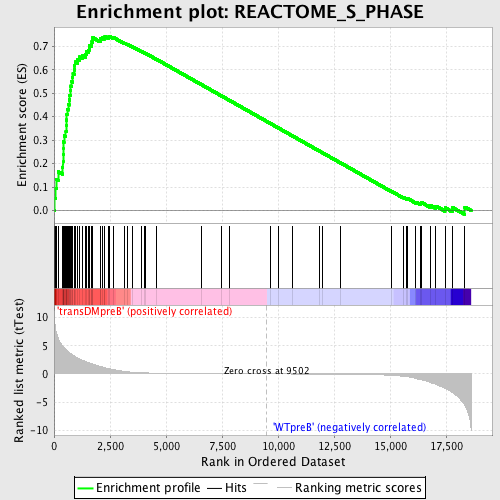
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | transDMpreB |
| GeneSet | REACTOME_S_PHASE |
| Enrichment Score (ES) | 0.74313015 |
| Normalized Enrichment Score (NES) | 1.7326553 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0022263236 |
| FWER p-Value | 0.02 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CCND1 | 4487 4488 8707 17535 | 25 | 9.348 | 0.0524 | Yes | ||
| 2 | POLD2 | 20537 | 78 | 7.693 | 0.0938 | Yes | ||
| 3 | POLE | 16755 | 111 | 7.095 | 0.1329 | Yes | ||
| 4 | PSMD8 | 7166 | 184 | 6.298 | 0.1653 | Yes | ||
| 5 | MCM3 | 13991 | 372 | 4.986 | 0.1838 | Yes | ||
| 6 | POLA2 | 23988 | 396 | 4.885 | 0.2107 | Yes | ||
| 7 | PSMA7 | 14318 | 403 | 4.860 | 0.2383 | Yes | ||
| 8 | CDC25A | 8721 | 411 | 4.833 | 0.2657 | Yes | ||
| 9 | PSMB9 | 23021 | 417 | 4.798 | 0.2931 | Yes | ||
| 10 | MCM2 | 17074 | 445 | 4.703 | 0.3186 | Yes | ||
| 11 | PSMD7 | 18752 3825 | 527 | 4.292 | 0.3390 | Yes | ||
| 12 | PSMC5 | 1348 20624 | 544 | 4.232 | 0.3624 | Yes | ||
| 13 | PSME3 | 20657 | 554 | 4.191 | 0.3860 | Yes | ||
| 14 | RFC5 | 13005 7791 | 568 | 4.143 | 0.4092 | Yes | ||
| 15 | PSMD3 | 5803 | 585 | 4.073 | 0.4317 | Yes | ||
| 16 | RPA1 | 20349 | 648 | 3.903 | 0.4508 | Yes | ||
| 17 | RFC4 | 1735 22627 | 670 | 3.827 | 0.4717 | Yes | ||
| 18 | RFC1 | 16527 | 696 | 3.717 | 0.4917 | Yes | ||
| 19 | PRIM1 | 19847 | 719 | 3.648 | 0.5115 | Yes | ||
| 20 | POLD3 | 17742 | 746 | 3.578 | 0.5307 | Yes | ||
| 21 | PSMA4 | 11179 | 787 | 3.478 | 0.5485 | Yes | ||
| 22 | PSMD12 | 20621 | 827 | 3.364 | 0.5658 | Yes | ||
| 23 | MCM5 | 18564 | 842 | 3.327 | 0.5842 | Yes | ||
| 24 | RPA2 | 2330 16057 | 895 | 3.168 | 0.5996 | Yes | ||
| 25 | FEN1 | 8961 | 915 | 3.126 | 0.6165 | Yes | ||
| 26 | RPA3 | 12667 | 932 | 3.065 | 0.6333 | Yes | ||
| 27 | PSME1 | 5300 9637 | 1039 | 2.818 | 0.6438 | Yes | ||
| 28 | PCNA | 9535 | 1127 | 2.676 | 0.6545 | Yes | ||
| 29 | PSMC4 | 2141 17915 | 1254 | 2.451 | 0.6618 | Yes | ||
| 30 | CUL1 | 17462 | 1412 | 2.216 | 0.6660 | Yes | ||
| 31 | CDK2 | 3438 3373 19592 3322 | 1447 | 2.143 | 0.6765 | Yes | ||
| 32 | MCM7 | 9372 3568 | 1539 | 1.995 | 0.6831 | Yes | ||
| 33 | MNAT1 | 9396 2161 | 1564 | 1.959 | 0.6931 | Yes | ||
| 34 | PSMB5 | 9633 | 1584 | 1.927 | 0.7031 | Yes | ||
| 35 | PSMC2 | 16909 | 1663 | 1.822 | 0.7094 | Yes | ||
| 36 | PSMD14 | 15005 | 1687 | 1.787 | 0.7184 | Yes | ||
| 37 | PSMB6 | 9634 | 1715 | 1.746 | 0.7270 | Yes | ||
| 38 | ORC5L | 11173 3595 | 1726 | 1.738 | 0.7365 | Yes | ||
| 39 | RFC3 | 12786 | 2067 | 1.318 | 0.7257 | Yes | ||
| 40 | PSMA5 | 6464 | 2069 | 1.318 | 0.7332 | Yes | ||
| 41 | CCNH | 7322 | 2152 | 1.209 | 0.7358 | Yes | ||
| 42 | PSMD9 | 3461 7393 | 2249 | 1.096 | 0.7369 | Yes | ||
| 43 | PSMD2 | 10137 5724 | 2266 | 1.069 | 0.7422 | Yes | ||
| 44 | PSMC1 | 9635 | 2420 | 0.965 | 0.7395 | Yes | ||
| 45 | PSMD10 | 2578 2597 24046 | 2453 | 0.936 | 0.7431 | Yes | ||
| 46 | PSMC6 | 7379 | 2641 | 0.793 | 0.7376 | No | ||
| 47 | PSMC3 | 9636 | 3137 | 0.443 | 0.7135 | No | ||
| 48 | PSMB3 | 11180 | 3281 | 0.380 | 0.7079 | No | ||
| 49 | WEE1 | 18127 | 3500 | 0.297 | 0.6979 | No | ||
| 50 | PSMD4 | 15251 | 3885 | 0.213 | 0.6784 | No | ||
| 51 | PSMB4 | 15252 | 4036 | 0.186 | 0.6714 | No | ||
| 52 | PSMA2 | 9631 | 4081 | 0.179 | 0.6700 | No | ||
| 53 | ORC4L | 11172 6460 | 4571 | 0.126 | 0.6444 | No | ||
| 54 | CDC45L | 22642 1752 | 6568 | 0.043 | 0.5370 | No | ||
| 55 | PSMD5 | 2774 14612 | 7473 | 0.027 | 0.4884 | No | ||
| 56 | POLE2 | 21053 | 7814 | 0.022 | 0.4702 | No | ||
| 57 | POLA1 | 24112 | 9653 | -0.002 | 0.3711 | No | ||
| 58 | CDK7 | 21365 | 10035 | -0.007 | 0.3506 | No | ||
| 59 | PSMA1 | 1627 17669 | 10653 | -0.015 | 0.3174 | No | ||
| 60 | PSMA6 | 21270 | 11867 | -0.033 | 0.2522 | No | ||
| 61 | PSMA3 | 9632 5298 | 11971 | -0.035 | 0.2468 | No | ||
| 62 | CDK4 | 3424 19859 | 12800 | -0.055 | 0.2025 | No | ||
| 63 | ORC2L | 385 13949 | 15082 | -0.255 | 0.0809 | No | ||
| 64 | PSMD11 | 12772 7600 | 15591 | -0.417 | 0.0559 | No | ||
| 65 | MCM6 | 4000 13845 4119 | 15750 | -0.489 | 0.0502 | No | ||
| 66 | ORC1L | 327 16144 | 15769 | -0.497 | 0.0521 | No | ||
| 67 | LIG1 | 18388 1749 1493 | 16146 | -0.805 | 0.0365 | No | ||
| 68 | MCM4 | 22655 1708 | 16371 | -1.005 | 0.0302 | No | ||
| 69 | PSMB1 | 23118 | 16410 | -1.038 | 0.0341 | No | ||
| 70 | PSMB2 | 2324 16078 | 16807 | -1.518 | 0.0215 | No | ||
| 71 | POLD1 | 17847 | 17042 | -1.851 | 0.0195 | No | ||
| 72 | PSMB10 | 5299 18761 | 17450 | -2.545 | 0.0122 | No | ||
| 73 | RB1 | 21754 | 17773 | -3.276 | 0.0137 | No | ||
| 74 | CDKN1A | 4511 8729 | 18334 | -5.522 | 0.0152 | No |