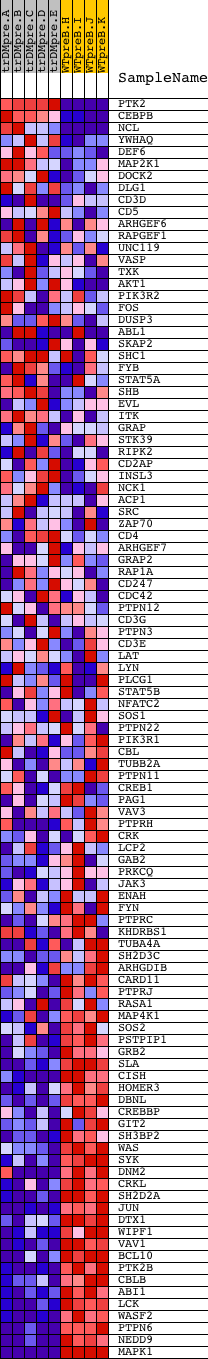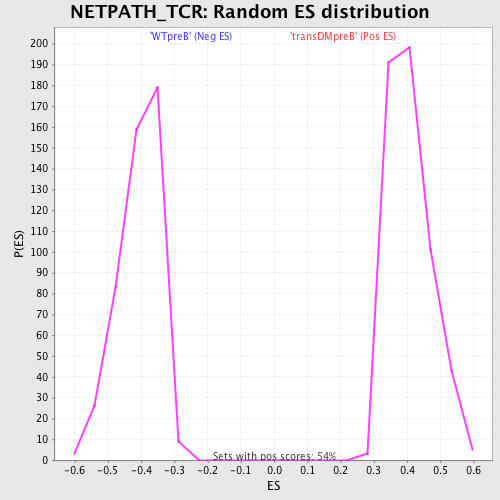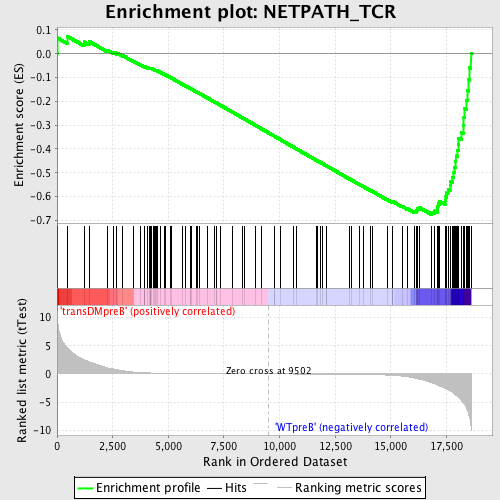
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | NETPATH_TCR |
| Enrichment Score (ES) | -0.67539734 |
| Normalized Enrichment Score (NES) | -1.6675596 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.031158853 |
| FWER p-Value | 0.203 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PTK2 | 22271 | 8 | 10.070 | 0.0668 | No | ||
| 2 | CEBPB | 8733 | 449 | 4.666 | 0.0742 | No | ||
| 3 | NCL | 5153 13899 | 1212 | 2.514 | 0.0498 | No | ||
| 4 | YWHAQ | 10369 | 1436 | 2.169 | 0.0522 | No | ||
| 5 | DEF6 | 23319 | 2251 | 1.091 | 0.0155 | No | ||
| 6 | MAP2K1 | 19082 | 2547 | 0.862 | 0.0054 | No | ||
| 7 | DOCK2 | 8243 | 2680 | 0.761 | 0.0033 | No | ||
| 8 | DLG1 | 22793 1672 | 2949 | 0.559 | -0.0074 | No | ||
| 9 | CD3D | 19473 | 3436 | 0.320 | -0.0316 | No | ||
| 10 | CD5 | 23741 | 3750 | 0.237 | -0.0469 | No | ||
| 11 | ARHGEF6 | 13297 13104 | 3918 | 0.207 | -0.0545 | No | ||
| 12 | RAPGEF1 | 4218 2860 | 3925 | 0.206 | -0.0535 | No | ||
| 13 | UNC119 | 20758 | 3934 | 0.203 | -0.0525 | No | ||
| 14 | VASP | 5847 | 4079 | 0.179 | -0.0591 | No | ||
| 15 | TXK | 16513 | 4155 | 0.170 | -0.0620 | No | ||
| 16 | AKT1 | 8568 | 4184 | 0.164 | -0.0625 | No | ||
| 17 | PIK3R2 | 18850 | 4208 | 0.163 | -0.0626 | No | ||
| 18 | FOS | 21202 | 4219 | 0.162 | -0.0621 | No | ||
| 19 | DUSP3 | 1358 13022 | 4241 | 0.159 | -0.0621 | No | ||
| 20 | ABL1 | 2693 4301 2794 | 4314 | 0.151 | -0.0650 | No | ||
| 21 | SKAP2 | 17153 | 4375 | 0.143 | -0.0673 | No | ||
| 22 | SHC1 | 9813 9812 5430 | 4421 | 0.139 | -0.0688 | No | ||
| 23 | FYB | 10717 | 4452 | 0.137 | -0.0695 | No | ||
| 24 | STAT5A | 20664 | 4510 | 0.131 | -0.0717 | No | ||
| 25 | SHB | 10493 | 4664 | 0.118 | -0.0792 | No | ||
| 26 | EVL | 21156 | 4820 | 0.108 | -0.0869 | No | ||
| 27 | ITK | 4934 | 4876 | 0.104 | -0.0891 | No | ||
| 28 | GRAP | 20851 | 5104 | 0.090 | -0.1008 | No | ||
| 29 | STK39 | 14570 | 5144 | 0.089 | -0.1023 | No | ||
| 30 | RIPK2 | 2528 15935 | 5624 | 0.067 | -0.1277 | No | ||
| 31 | CD2AP | 22975 | 5774 | 0.062 | -0.1354 | No | ||
| 32 | INSL3 | 9181 | 5980 | 0.057 | -0.1461 | No | ||
| 33 | NCK1 | 9447 5152 | 5996 | 0.056 | -0.1465 | No | ||
| 34 | ACP1 | 4317 | 6039 | 0.055 | -0.1484 | No | ||
| 35 | SRC | 5507 | 6271 | 0.049 | -0.1605 | No | ||
| 36 | ZAP70 | 14271 4042 | 6321 | 0.048 | -0.1629 | No | ||
| 37 | CD4 | 16999 | 6410 | 0.046 | -0.1673 | No | ||
| 38 | ARHGEF7 | 3823 | 6776 | 0.039 | -0.1868 | No | ||
| 39 | GRAP2 | 5113 9398 | 7076 | 0.033 | -0.2027 | No | ||
| 40 | RAP1A | 8467 | 7142 | 0.032 | -0.2060 | No | ||
| 41 | CD247 | 4498 8715 | 7180 | 0.032 | -0.2078 | No | ||
| 42 | CDC42 | 4503 8722 4504 2465 | 7363 | 0.028 | -0.2174 | No | ||
| 43 | PTPN12 | 16597 3502 3586 3665 | 7872 | 0.021 | -0.2447 | No | ||
| 44 | CD3G | 19139 | 8313 | 0.015 | -0.2684 | No | ||
| 45 | PTPN3 | 9661 | 8441 | 0.014 | -0.2752 | No | ||
| 46 | CD3E | 8714 | 8903 | 0.008 | -0.3000 | No | ||
| 47 | LAT | 17643 | 9183 | 0.004 | -0.3151 | No | ||
| 48 | LYN | 16281 | 9750 | -0.003 | -0.3456 | No | ||
| 49 | PLCG1 | 14753 | 9775 | -0.004 | -0.3469 | No | ||
| 50 | STAT5B | 20222 | 10037 | -0.007 | -0.3609 | No | ||
| 51 | NFATC2 | 5168 2866 | 10619 | -0.014 | -0.3922 | No | ||
| 52 | SOS1 | 5476 | 10751 | -0.016 | -0.3992 | No | ||
| 53 | PTPN22 | 1775 15470 | 11643 | -0.030 | -0.4471 | No | ||
| 54 | PIK3R1 | 3170 | 11694 | -0.031 | -0.4496 | No | ||
| 55 | CBL | 19154 | 11843 | -0.033 | -0.4574 | No | ||
| 56 | TUBB2A | 21493 | 11913 | -0.034 | -0.4609 | No | ||
| 57 | PTPN11 | 5326 16391 9660 | 12087 | -0.038 | -0.4700 | No | ||
| 58 | CREB1 | 3990 8782 4558 4093 | 12096 | -0.038 | -0.4702 | No | ||
| 59 | PAG1 | 15376 | 13138 | -0.066 | -0.5260 | No | ||
| 60 | VAV3 | 1774 1848 15443 | 13236 | -0.069 | -0.5307 | No | ||
| 61 | PTPRH | 11861 | 13593 | -0.085 | -0.5494 | No | ||
| 62 | CRK | 4559 1249 | 13780 | -0.096 | -0.5588 | No | ||
| 63 | LCP2 | 4988 9268 | 14104 | -0.118 | -0.5755 | No | ||
| 64 | GAB2 | 1821 18184 2025 | 14159 | -0.123 | -0.5776 | No | ||
| 65 | PRKCQ | 2873 2831 | 14839 | -0.204 | -0.6129 | No | ||
| 66 | JAK3 | 9198 4936 | 15055 | -0.249 | -0.6228 | No | ||
| 67 | ENAH | 4665 8901 | 15070 | -0.252 | -0.6219 | No | ||
| 68 | FYN | 3375 3395 20052 | 15076 | -0.254 | -0.6205 | No | ||
| 69 | PTPRC | 5327 9662 | 15502 | -0.380 | -0.6409 | No | ||
| 70 | KHDRBS1 | 5405 9778 | 15739 | -0.484 | -0.6504 | No | ||
| 71 | TUBA4A | 10232 | 16060 | -0.705 | -0.6630 | No | ||
| 72 | SH2D3C | 2864 | 16139 | -0.792 | -0.6619 | No | ||
| 73 | ARHGDIB | 16950 | 16142 | -0.803 | -0.6567 | No | ||
| 74 | CARD11 | 8436 | 16201 | -0.863 | -0.6540 | No | ||
| 75 | PTPRJ | 9664 | 16204 | -0.867 | -0.6484 | No | ||
| 76 | RASA1 | 10174 | 16303 | -0.967 | -0.6472 | No | ||
| 77 | MAP4K1 | 18313 | 16826 | -1.546 | -0.6651 | Yes | ||
| 78 | SOS2 | 21049 | 16958 | -1.731 | -0.6606 | Yes | ||
| 79 | PSTPIP1 | 19437 | 17089 | -1.941 | -0.6547 | Yes | ||
| 80 | GRB2 | 20149 | 17099 | -1.953 | -0.6421 | Yes | ||
| 81 | SLA | 5449 | 17124 | -1.995 | -0.6301 | Yes | ||
| 82 | CISH | 8743 | 17180 | -2.104 | -0.6190 | Yes | ||
| 83 | HOMER3 | 18590 | 17457 | -2.554 | -0.6169 | Yes | ||
| 84 | DBNL | 20954 | 17476 | -2.584 | -0.6006 | Yes | ||
| 85 | CREBBP | 22682 8783 | 17516 | -2.659 | -0.5849 | Yes | ||
| 86 | GIT2 | 11174 6462 | 17572 | -2.781 | -0.5694 | Yes | ||
| 87 | SH3BP2 | 16874 | 17663 | -2.971 | -0.5544 | Yes | ||
| 88 | WAS | 24193 | 17688 | -3.040 | -0.5354 | Yes | ||
| 89 | SYK | 21636 | 17785 | -3.326 | -0.5184 | Yes | ||
| 90 | DNM2 | 4635 3103 | 17822 | -3.423 | -0.4975 | Yes | ||
| 91 | CRKL | 4560 | 17866 | -3.568 | -0.4760 | Yes | ||
| 92 | SH2D2A | 6569 | 17915 | -3.712 | -0.4538 | Yes | ||
| 93 | JUN | 15832 | 17929 | -3.751 | -0.4294 | Yes | ||
| 94 | DTX1 | 16396 | 17992 | -3.927 | -0.4066 | Yes | ||
| 95 | WIPF1 | 14560 | 18027 | -4.039 | -0.3814 | Yes | ||
| 96 | VAV1 | 23173 | 18053 | -4.129 | -0.3552 | Yes | ||
| 97 | BCL10 | 15397 | 18173 | -4.717 | -0.3302 | Yes | ||
| 98 | PTK2B | 21776 | 18273 | -5.239 | -0.3005 | Yes | ||
| 99 | CBLB | 5531 22734 | 18286 | -5.302 | -0.2658 | Yes | ||
| 100 | ABI1 | 4300 | 18324 | -5.473 | -0.2313 | Yes | ||
| 101 | LCK | 15746 | 18405 | -6.029 | -0.1953 | Yes | ||
| 102 | WASF2 | 6326 | 18455 | -6.597 | -0.1539 | Yes | ||
| 103 | PTPN6 | 17002 | 18509 | -7.257 | -0.1083 | Yes | ||
| 104 | NEDD9 | 3206 21483 | 18547 | -7.820 | -0.0581 | Yes | ||
| 105 | MAPK1 | 1642 11167 | 18603 | -9.259 | 0.0007 | Yes |