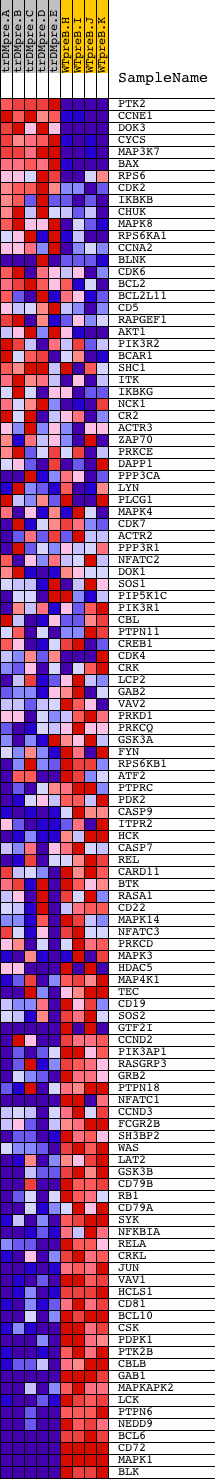
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
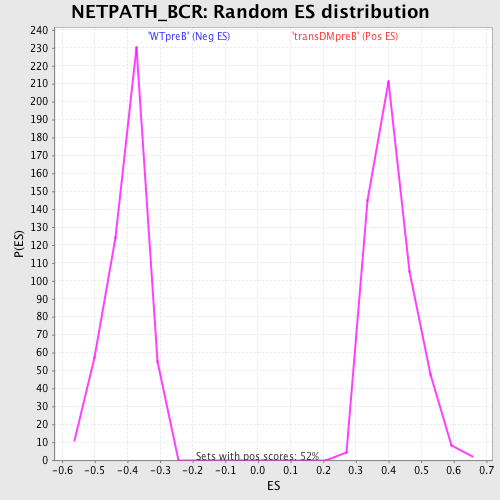
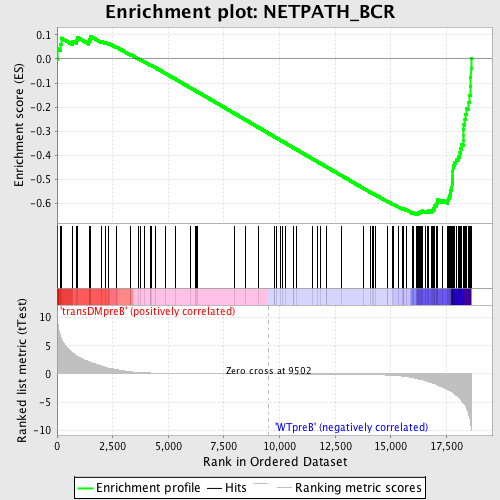
| GeneSet | NETPATH_BCR |
| Enrichment Score (ES) | -0.6449906 |
| Normalized Enrichment Score (NES) | -1.6132458 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.04336412 |
| FWER p-Value | 0.492 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PTK2 | 22271 | 8 | 10.070 | 0.0420 | No | ||
| 2 | CCNE1 | 17857 | 159 | 6.569 | 0.0615 | No | ||
| 3 | DOK3 | 3229 21451 | 182 | 6.330 | 0.0870 | No | ||
| 4 | CYCS | 8821 | 708 | 3.673 | 0.0740 | No | ||
| 5 | MAP3K7 | 16255 | 878 | 3.198 | 0.0784 | No | ||
| 6 | BAX | 17832 | 898 | 3.158 | 0.0906 | No | ||
| 7 | RPS6 | 9757 | 1433 | 2.175 | 0.0709 | No | ||
| 8 | CDK2 | 3438 3373 19592 3322 | 1447 | 2.143 | 0.0792 | No | ||
| 9 | IKBKB | 4907 | 1483 | 2.082 | 0.0861 | No | ||
| 10 | CHUK | 23665 | 1487 | 2.079 | 0.0947 | No | ||
| 11 | MAPK8 | 6459 | 2014 | 1.381 | 0.0721 | No | ||
| 12 | RPS6KA1 | 15725 | 2164 | 1.195 | 0.0691 | No | ||
| 13 | CCNA2 | 15357 | 2326 | 1.015 | 0.0646 | No | ||
| 14 | BLNK | 23681 3691 | 2670 | 0.771 | 0.0493 | No | ||
| 15 | CDK6 | 16600 | 3298 | 0.374 | 0.0170 | No | ||
| 16 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 3317 | 0.365 | 0.0176 | No | ||
| 17 | BCL2L11 | 2790 14861 | 3657 | 0.259 | 0.0004 | No | ||
| 18 | CD5 | 23741 | 3750 | 0.237 | -0.0036 | No | ||
| 19 | RAPGEF1 | 4218 2860 | 3925 | 0.206 | -0.0121 | No | ||
| 20 | AKT1 | 8568 | 4184 | 0.164 | -0.0254 | No | ||
| 21 | PIK3R2 | 18850 | 4208 | 0.163 | -0.0260 | No | ||
| 22 | BCAR1 | 18741 | 4251 | 0.158 | -0.0276 | No | ||
| 23 | SHC1 | 9813 9812 5430 | 4421 | 0.139 | -0.0361 | No | ||
| 24 | ITK | 4934 | 4876 | 0.104 | -0.0602 | No | ||
| 25 | IKBKG | 2570 2562 4908 | 5302 | 0.081 | -0.0828 | No | ||
| 26 | NCK1 | 9447 5152 | 5996 | 0.056 | -0.1201 | No | ||
| 27 | CR2 | 3942 13707 | 6213 | 0.051 | -0.1315 | No | ||
| 28 | ACTR3 | 13166 | 6253 | 0.049 | -0.1334 | No | ||
| 29 | ZAP70 | 14271 4042 | 6321 | 0.048 | -0.1368 | No | ||
| 30 | PRKCE | 9575 | 7969 | 0.020 | -0.2258 | No | ||
| 31 | DAPP1 | 11159 6445 6446 | 8472 | 0.013 | -0.2529 | No | ||
| 32 | PPP3CA | 1863 5284 | 9030 | 0.006 | -0.2829 | No | ||
| 33 | LYN | 16281 | 9750 | -0.003 | -0.3218 | No | ||
| 34 | PLCG1 | 14753 | 9775 | -0.004 | -0.3231 | No | ||
| 35 | MAPK4 | 23409 | 9852 | -0.005 | -0.3271 | No | ||
| 36 | CDK7 | 21365 | 10035 | -0.007 | -0.3370 | No | ||
| 37 | ACTR2 | 20523 | 10132 | -0.008 | -0.3421 | No | ||
| 38 | PPP3R1 | 9611 489 | 10273 | -0.010 | -0.3496 | No | ||
| 39 | NFATC2 | 5168 2866 | 10619 | -0.014 | -0.3682 | No | ||
| 40 | DOK1 | 17104 1018 1177 | 10626 | -0.015 | -0.3685 | No | ||
| 41 | SOS1 | 5476 | 10751 | -0.016 | -0.3751 | No | ||
| 42 | PIP5K1C | 5250 19929 | 11474 | -0.027 | -0.4140 | No | ||
| 43 | PIK3R1 | 3170 | 11694 | -0.031 | -0.4257 | No | ||
| 44 | CBL | 19154 | 11843 | -0.033 | -0.4336 | No | ||
| 45 | PTPN11 | 5326 16391 9660 | 12087 | -0.038 | -0.4466 | No | ||
| 46 | CREB1 | 3990 8782 4558 4093 | 12096 | -0.038 | -0.4468 | No | ||
| 47 | CDK4 | 3424 19859 | 12800 | -0.055 | -0.4846 | No | ||
| 48 | CRK | 4559 1249 | 13780 | -0.096 | -0.5371 | No | ||
| 49 | LCP2 | 4988 9268 | 14104 | -0.118 | -0.5541 | No | ||
| 50 | GAB2 | 1821 18184 2025 | 14159 | -0.123 | -0.5565 | No | ||
| 51 | VAV2 | 5848 2670 | 14239 | -0.130 | -0.5602 | No | ||
| 52 | PRKD1 | 2074 21075 | 14288 | -0.135 | -0.5622 | No | ||
| 53 | PRKCQ | 2873 2831 | 14839 | -0.204 | -0.5911 | No | ||
| 54 | GSK3A | 405 | 14869 | -0.210 | -0.5918 | No | ||
| 55 | FYN | 3375 3395 20052 | 15076 | -0.254 | -0.6018 | No | ||
| 56 | RPS6KB1 | 7815 1207 13040 | 15117 | -0.261 | -0.6029 | No | ||
| 57 | ATF2 | 4418 2759 | 15353 | -0.322 | -0.6142 | No | ||
| 58 | PTPRC | 5327 9662 | 15502 | -0.380 | -0.6206 | No | ||
| 59 | PDK2 | 20285 | 15551 | -0.404 | -0.6215 | No | ||
| 60 | CASP9 | 16001 2410 2458 | 15567 | -0.410 | -0.6206 | No | ||
| 61 | ITPR2 | 9194 | 15707 | -0.466 | -0.6262 | No | ||
| 62 | HCK | 14787 | 15955 | -0.616 | -0.6369 | No | ||
| 63 | CASP7 | 23826 | 16022 | -0.678 | -0.6376 | No | ||
| 64 | REL | 9716 | 16159 | -0.818 | -0.6415 | Yes | ||
| 65 | CARD11 | 8436 | 16201 | -0.863 | -0.6401 | Yes | ||
| 66 | BTK | 24061 | 16235 | -0.895 | -0.6381 | Yes | ||
| 67 | RASA1 | 10174 | 16303 | -0.967 | -0.6377 | Yes | ||
| 68 | CD22 | 17881 | 16316 | -0.978 | -0.6342 | Yes | ||
| 69 | MAPK14 | 23313 | 16395 | -1.021 | -0.6341 | Yes | ||
| 70 | NFATC3 | 5169 | 16404 | -1.032 | -0.6302 | Yes | ||
| 71 | PRKCD | 21897 | 16559 | -1.188 | -0.6336 | Yes | ||
| 72 | MAPK3 | 6458 11170 | 16645 | -1.295 | -0.6327 | Yes | ||
| 73 | HDAC5 | 1480 20205 | 16676 | -1.338 | -0.6287 | Yes | ||
| 74 | MAP4K1 | 18313 | 16826 | -1.546 | -0.6302 | Yes | ||
| 75 | TEC | 16514 | 16891 | -1.651 | -0.6267 | Yes | ||
| 76 | CD19 | 17640 | 16921 | -1.697 | -0.6212 | Yes | ||
| 77 | SOS2 | 21049 | 16958 | -1.731 | -0.6158 | Yes | ||
| 78 | GTF2I | 16353 5491 9863 3280 3184 | 16975 | -1.748 | -0.6093 | Yes | ||
| 79 | CCND2 | 16987 | 17040 | -1.850 | -0.6050 | Yes | ||
| 80 | PIK3AP1 | 23679 | 17057 | -1.885 | -0.5979 | Yes | ||
| 81 | RASGRP3 | 10767 | 17096 | -1.946 | -0.5918 | Yes | ||
| 82 | GRB2 | 20149 | 17099 | -1.953 | -0.5837 | Yes | ||
| 83 | PTPN18 | 14279 | 17310 | -2.320 | -0.5853 | Yes | ||
| 84 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 17547 | -2.734 | -0.5865 | Yes | ||
| 85 | CCND3 | 4489 4490 | 17589 | -2.825 | -0.5768 | Yes | ||
| 86 | FCGR2B | 8959 | 17622 | -2.880 | -0.5665 | Yes | ||
| 87 | SH3BP2 | 16874 | 17663 | -2.971 | -0.5561 | Yes | ||
| 88 | WAS | 24193 | 17688 | -3.040 | -0.5446 | Yes | ||
| 89 | LAT2 | 16351 | 17736 | -3.151 | -0.5339 | Yes | ||
| 90 | GSK3B | 22761 | 17749 | -3.186 | -0.5211 | Yes | ||
| 91 | CD79B | 20185 1309 | 17759 | -3.226 | -0.5080 | Yes | ||
| 92 | RB1 | 21754 | 17773 | -3.276 | -0.4949 | Yes | ||
| 93 | CD79A | 18342 | 17781 | -3.312 | -0.4814 | Yes | ||
| 94 | SYK | 21636 | 17785 | -3.326 | -0.4675 | Yes | ||
| 95 | NFKBIA | 21065 | 17794 | -3.356 | -0.4539 | Yes | ||
| 96 | RELA | 23783 | 17808 | -3.389 | -0.4403 | Yes | ||
| 97 | CRKL | 4560 | 17866 | -3.568 | -0.4284 | Yes | ||
| 98 | JUN | 15832 | 17929 | -3.751 | -0.4159 | Yes | ||
| 99 | VAV1 | 23173 | 18053 | -4.129 | -0.4052 | Yes | ||
| 100 | HCLS1 | 22770 | 18103 | -4.347 | -0.3895 | Yes | ||
| 101 | CD81 | 8719 | 18143 | -4.545 | -0.3725 | Yes | ||
| 102 | BCL10 | 15397 | 18173 | -4.717 | -0.3542 | Yes | ||
| 103 | CSK | 8805 | 18265 | -5.201 | -0.3372 | Yes | ||
| 104 | PDPK1 | 23097 | 18270 | -5.228 | -0.3155 | Yes | ||
| 105 | PTK2B | 21776 | 18273 | -5.239 | -0.2935 | Yes | ||
| 106 | CBLB | 5531 22734 | 18286 | -5.302 | -0.2718 | Yes | ||
| 107 | GAB1 | 18828 | 18330 | -5.503 | -0.2510 | Yes | ||
| 108 | MAPKAPK2 | 13838 | 18361 | -5.702 | -0.2286 | Yes | ||
| 109 | LCK | 15746 | 18405 | -6.029 | -0.2056 | Yes | ||
| 110 | PTPN6 | 17002 | 18509 | -7.257 | -0.1806 | Yes | ||
| 111 | NEDD9 | 3206 21483 | 18547 | -7.820 | -0.1497 | Yes | ||
| 112 | BCL6 | 22624 | 18584 | -8.751 | -0.1148 | Yes | ||
| 113 | CD72 | 8718 | 18591 | -8.983 | -0.0773 | Yes | ||
| 114 | MAPK1 | 1642 11167 | 18603 | -9.259 | -0.0389 | Yes | ||
| 115 | BLK | 3205 21791 | 18607 | -9.402 | 0.0005 | Yes |