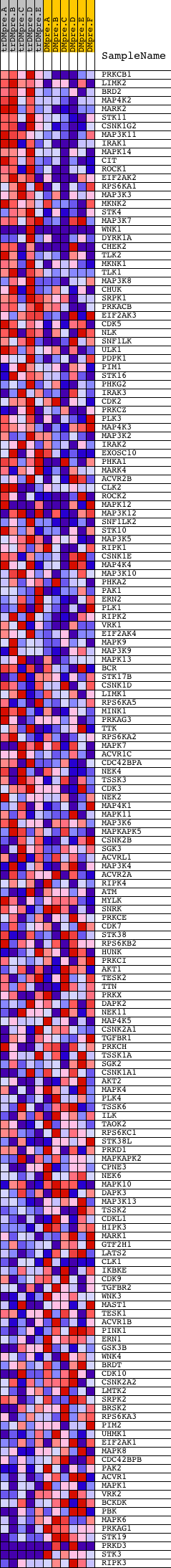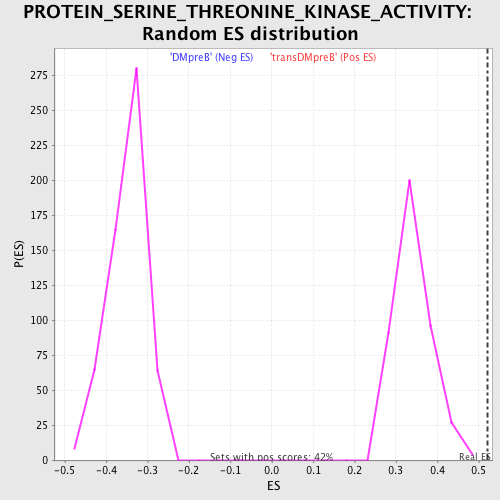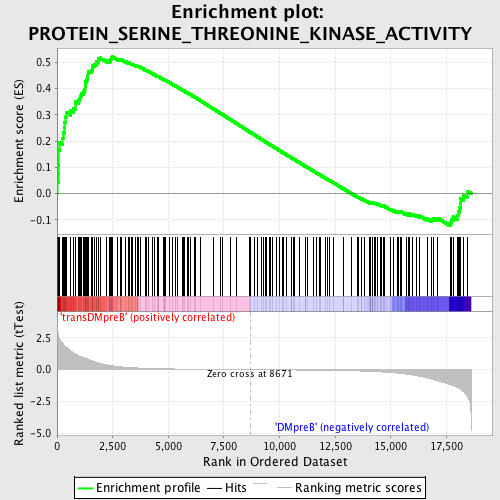
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | PROTEIN_SERINE_THREONINE_KINASE_ACTIVITY |
| Enrichment Score (ES) | 0.52048784 |
| Normalized Enrichment Score (NES) | 1.5221715 |
| Nominal p-value | 0.0 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PRKCB1 | 1693 9574 | 2 | 3.865 | 0.0450 | Yes | ||
| 2 | LIMK2 | 9278 1296 | 47 | 2.806 | 0.0754 | Yes | ||
| 3 | BRD2 | 4735 8984 7628 | 51 | 2.773 | 0.1076 | Yes | ||
| 4 | MAP4K2 | 6457 | 72 | 2.603 | 0.1369 | Yes | ||
| 5 | MARK2 | 8899 | 74 | 2.595 | 0.1672 | Yes | ||
| 6 | STK11 | 19950 | 114 | 2.391 | 0.1930 | Yes | ||
| 7 | CSNK1G2 | 19940 3418 | 246 | 2.062 | 0.2100 | Yes | ||
| 8 | MAP3K11 | 11163 | 275 | 2.001 | 0.2318 | Yes | ||
| 9 | IRAK1 | 4916 | 322 | 1.872 | 0.2512 | Yes | ||
| 10 | MAPK14 | 23313 | 334 | 1.857 | 0.2723 | Yes | ||
| 11 | CIT | 16727 | 367 | 1.809 | 0.2917 | Yes | ||
| 12 | ROCK1 | 5386 | 409 | 1.746 | 0.3098 | Yes | ||
| 13 | EIF2AK2 | 22892 | 621 | 1.457 | 0.3154 | Yes | ||
| 14 | RPS6KA1 | 15725 | 736 | 1.314 | 0.3246 | Yes | ||
| 15 | MAP3K3 | 20626 | 808 | 1.254 | 0.3354 | Yes | ||
| 16 | MKNK2 | 3299 9392 | 809 | 1.250 | 0.3500 | Yes | ||
| 17 | STK4 | 14743 | 965 | 1.084 | 0.3542 | Yes | ||
| 18 | MAP3K7 | 16255 | 1018 | 1.035 | 0.3635 | Yes | ||
| 19 | WNK1 | 1109 17020 6106 | 1071 | 1.007 | 0.3724 | Yes | ||
| 20 | DYRK1A | 4649 | 1113 | 0.993 | 0.3818 | Yes | ||
| 21 | CHEK2 | 16751 3587 | 1190 | 0.938 | 0.3886 | Yes | ||
| 22 | TLK2 | 20628 | 1238 | 0.905 | 0.3966 | Yes | ||
| 23 | MKNK1 | 2504 | 1254 | 0.891 | 0.4062 | Yes | ||
| 24 | TLK1 | 14563 | 1260 | 0.887 | 0.4163 | Yes | ||
| 25 | MAP3K8 | 23495 | 1281 | 0.872 | 0.4254 | Yes | ||
| 26 | CHUK | 23665 | 1310 | 0.853 | 0.4339 | Yes | ||
| 27 | SRPK1 | 23049 | 1373 | 0.817 | 0.4400 | Yes | ||
| 28 | PRKACB | 15140 | 1381 | 0.813 | 0.4492 | Yes | ||
| 29 | EIF2AK3 | 17421 | 1388 | 0.809 | 0.4583 | Yes | ||
| 30 | CDK5 | 16591 | 1415 | 0.791 | 0.4661 | Yes | ||
| 31 | NLK | 5179 5178 | 1529 | 0.723 | 0.4684 | Yes | ||
| 32 | SNF1LK | 23033 | 1570 | 0.699 | 0.4744 | Yes | ||
| 33 | ULK1 | 16436 | 1603 | 0.681 | 0.4806 | Yes | ||
| 34 | PDPK1 | 23097 | 1610 | 0.675 | 0.4882 | Yes | ||
| 35 | PIM1 | 23308 | 1691 | 0.635 | 0.4912 | Yes | ||
| 36 | STK16 | 14216 | 1753 | 0.596 | 0.4949 | Yes | ||
| 37 | PHKG2 | 18065 | 1764 | 0.591 | 0.5013 | Yes | ||
| 38 | IRAK3 | 19617 | 1842 | 0.541 | 0.5034 | Yes | ||
| 39 | CDK2 | 3438 3373 19592 3322 | 1865 | 0.529 | 0.5084 | Yes | ||
| 40 | PRKCZ | 5260 | 1877 | 0.520 | 0.5139 | Yes | ||
| 41 | PLK3 | 15790 | 1938 | 0.487 | 0.5163 | Yes | ||
| 42 | MAP4K3 | 22887 | 2224 | 0.377 | 0.5052 | Yes | ||
| 43 | MAP3K2 | 11165 | 2345 | 0.340 | 0.5027 | Yes | ||
| 44 | IRAK2 | 17324 | 2364 | 0.332 | 0.5056 | Yes | ||
| 45 | EXOSC10 | 2482 11943 | 2402 | 0.322 | 0.5074 | Yes | ||
| 46 | PHKA1 | 2645 24089 2582 | 2405 | 0.321 | 0.5110 | Yes | ||
| 47 | MARK4 | 10572 | 2427 | 0.313 | 0.5135 | Yes | ||
| 48 | ACVR2B | 19276 | 2436 | 0.310 | 0.5167 | Yes | ||
| 49 | CLK2 | 1883 15544 | 2477 | 0.298 | 0.5180 | Yes | ||
| 50 | ROCK2 | 21309 5387 | 2496 | 0.295 | 0.5205 | Yes | ||
| 51 | MAPK12 | 2182 2215 22164 | 2721 | 0.246 | 0.5112 | No | ||
| 52 | MAP3K12 | 6454 11164 | 2832 | 0.229 | 0.5079 | No | ||
| 53 | SNF1LK2 | 6168 10649 | 2838 | 0.228 | 0.5103 | No | ||
| 54 | STK10 | 5532 9916 | 2897 | 0.217 | 0.5097 | No | ||
| 55 | MAP3K5 | 20079 11166 | 3069 | 0.191 | 0.5027 | No | ||
| 56 | RIPK1 | 5381 | 3213 | 0.170 | 0.4969 | No | ||
| 57 | CSNK1E | 6570 2211 11332 | 3242 | 0.166 | 0.4973 | No | ||
| 58 | MAP4K4 | 11241 | 3363 | 0.151 | 0.4926 | No | ||
| 59 | MAP3K10 | 3831 17916 | 3402 | 0.147 | 0.4922 | No | ||
| 60 | PHKA2 | 8471 24216 | 3539 | 0.135 | 0.4864 | No | ||
| 61 | PAK1 | 9527 | 3601 | 0.130 | 0.4846 | No | ||
| 62 | ERN2 | 17651 | 3631 | 0.128 | 0.4846 | No | ||
| 63 | PLK1 | 9590 5266 | 3640 | 0.127 | 0.4856 | No | ||
| 64 | RIPK2 | 2528 15935 | 3734 | 0.119 | 0.4820 | No | ||
| 65 | VRK1 | 5862 2049 | 3991 | 0.103 | 0.4693 | No | ||
| 66 | EIF2AK4 | 14909 | 4029 | 0.100 | 0.4685 | No | ||
| 67 | MAPK9 | 1233 20903 1383 | 4094 | 0.096 | 0.4661 | No | ||
| 68 | MAP3K9 | 6872 | 4297 | 0.084 | 0.4561 | No | ||
| 69 | MAPK13 | 23314 | 4397 | 0.080 | 0.4517 | No | ||
| 70 | BCR | 8478 3384 19990 | 4523 | 0.075 | 0.4458 | No | ||
| 71 | STK17B | 13957 | 4545 | 0.074 | 0.4455 | No | ||
| 72 | CSNK1D | 4181 8352 1387 | 4572 | 0.073 | 0.4450 | No | ||
| 73 | LIMK1 | 16350 | 4789 | 0.065 | 0.4340 | No | ||
| 74 | RPS6KA5 | 2076 21005 | 4837 | 0.064 | 0.4322 | No | ||
| 75 | MINK1 | 20802 | 4840 | 0.064 | 0.4328 | No | ||
| 76 | PRKAG3 | 6295 | 4882 | 0.063 | 0.4314 | No | ||
| 77 | TTK | 19366 | 5063 | 0.057 | 0.4223 | No | ||
| 78 | RPS6KA2 | 9759 9758 | 5200 | 0.053 | 0.4155 | No | ||
| 79 | MAPK7 | 1381 20414 | 5307 | 0.050 | 0.4104 | No | ||
| 80 | ACVR1C | 14585 | 5425 | 0.048 | 0.4046 | No | ||
| 81 | CDC42BPA | 10384 | 5645 | 0.043 | 0.3932 | No | ||
| 82 | NEK4 | 22061 3687 5534 | 5659 | 0.042 | 0.3930 | No | ||
| 83 | TSSK3 | 4388 | 5706 | 0.042 | 0.3910 | No | ||
| 84 | CDK3 | 20588 | 5843 | 0.039 | 0.3840 | No | ||
| 85 | NEK2 | 5160 | 5898 | 0.038 | 0.3816 | No | ||
| 86 | MAP4K1 | 18313 | 6010 | 0.036 | 0.3760 | No | ||
| 87 | MAPK11 | 2264 9618 22163 | 6170 | 0.033 | 0.3677 | No | ||
| 88 | MAP3K6 | 16054 | 6199 | 0.033 | 0.3666 | No | ||
| 89 | MAPKAPK5 | 16387 | 6436 | 0.029 | 0.3541 | No | ||
| 90 | CSNK2B | 23008 | 7009 | 0.020 | 0.3233 | No | ||
| 91 | SGK3 | 5025 4053 9312 3997 4025 | 7322 | 0.016 | 0.3066 | No | ||
| 92 | ACVRL1 | 22354 | 7450 | 0.014 | 0.2999 | No | ||
| 93 | MAP3K4 | 23126 | 7801 | 0.010 | 0.2810 | No | ||
| 94 | ACVR2A | 8542 | 8040 | 0.007 | 0.2682 | No | ||
| 95 | RIPK4 | 22526 | 8667 | 0.000 | 0.2343 | No | ||
| 96 | ATM | 2976 19115 | 8700 | -0.000 | 0.2325 | No | ||
| 97 | MYLK | 22778 4213 | 8871 | -0.002 | 0.2233 | No | ||
| 98 | SNRK | 5467 5468 9840 | 9022 | -0.004 | 0.2153 | No | ||
| 99 | PRKCE | 9575 | 9170 | -0.006 | 0.2074 | No | ||
| 100 | CDK7 | 21365 | 9267 | -0.007 | 0.2022 | No | ||
| 101 | STK38 | 23045 | 9365 | -0.009 | 0.1971 | No | ||
| 102 | RPS6KB2 | 7198 | 9416 | -0.009 | 0.1945 | No | ||
| 103 | HUNK | 22708 | 9526 | -0.011 | 0.1887 | No | ||
| 104 | PRKCI | 9576 | 9605 | -0.012 | 0.1846 | No | ||
| 105 | AKT1 | 8568 | 9662 | -0.013 | 0.1817 | No | ||
| 106 | TESK2 | 10504 | 9879 | -0.015 | 0.1702 | No | ||
| 107 | TTN | 14552 2743 14553 | 9974 | -0.016 | 0.1653 | No | ||
| 108 | PRKX | 9619 5290 | 10149 | -0.019 | 0.1561 | No | ||
| 109 | DAPK2 | 19395 | 10195 | -0.019 | 0.1538 | No | ||
| 110 | NEK11 | 3099 19016 | 10302 | -0.021 | 0.1483 | No | ||
| 111 | MAP4K5 | 11853 21048 2101 2142 | 10536 | -0.024 | 0.1360 | No | ||
| 112 | CSNK2A1 | 14797 | 10544 | -0.024 | 0.1359 | No | ||
| 113 | TGFBR1 | 16213 10165 5745 | 10621 | -0.025 | 0.1321 | No | ||
| 114 | PRKCH | 21246 | 10685 | -0.026 | 0.1290 | No | ||
| 115 | TSSK1A | 5802 | 10898 | -0.029 | 0.1178 | No | ||
| 116 | SGK2 | 14750 | 11144 | -0.033 | 0.1049 | No | ||
| 117 | CSNK1A1 | 8204 | 11168 | -0.034 | 0.1041 | No | ||
| 118 | AKT2 | 4365 4366 | 11247 | -0.035 | 0.1002 | No | ||
| 119 | MAPK4 | 23409 | 11523 | -0.039 | 0.0858 | No | ||
| 120 | PLK4 | 1870 1874 | 11649 | -0.042 | 0.0795 | No | ||
| 121 | TSSK6 | 18594 | 11786 | -0.045 | 0.0726 | No | ||
| 122 | ILK | 9177 1618 | 11833 | -0.046 | 0.0707 | No | ||
| 123 | TAOK2 | 10610 3794 | 12072 | -0.051 | 0.0584 | No | ||
| 124 | RPS6KC1 | 6729 | 12158 | -0.053 | 0.0544 | No | ||
| 125 | STK38L | 17240 | 12247 | -0.055 | 0.0502 | No | ||
| 126 | PRKD1 | 2074 21075 | 12420 | -0.059 | 0.0416 | No | ||
| 127 | MAPKAPK2 | 13838 | 12872 | -0.073 | 0.0180 | No | ||
| 128 | CPNE3 | 7689 | 12878 | -0.073 | 0.0186 | No | ||
| 129 | NEK6 | 15018 | 13233 | -0.087 | 0.0004 | No | ||
| 130 | MAPK10 | 11169 | 13511 | -0.099 | -0.0134 | No | ||
| 131 | DAPK3 | 19932 | 13525 | -0.100 | -0.0130 | No | ||
| 132 | MAP3K13 | 22814 | 13677 | -0.108 | -0.0199 | No | ||
| 133 | TSSK2 | 10225 | 13695 | -0.109 | -0.0196 | No | ||
| 134 | CDKL1 | 12914 | 13816 | -0.116 | -0.0247 | No | ||
| 135 | HIPK3 | 14504 | 14019 | -0.129 | -0.0342 | No | ||
| 136 | MARK1 | 5955 | 14063 | -0.131 | -0.0350 | No | ||
| 137 | GTF2H1 | 4069 18236 | 14080 | -0.132 | -0.0343 | No | ||
| 138 | LATS2 | 11923 6928 | 14097 | -0.134 | -0.0336 | No | ||
| 139 | CLK1 | 13951 4127 4529 8748 | 14160 | -0.138 | -0.0353 | No | ||
| 140 | IKBKE | 67 | 14176 | -0.139 | -0.0345 | No | ||
| 141 | CDK9 | 14619 | 14177 | -0.139 | -0.0329 | No | ||
| 142 | TGFBR2 | 2994 3001 10167 | 14270 | -0.147 | -0.0362 | No | ||
| 143 | WNK3 | 24227 | 14319 | -0.151 | -0.0370 | No | ||
| 144 | MAST1 | 12143 | 14378 | -0.157 | -0.0383 | No | ||
| 145 | TESK1 | 16233 | 14514 | -0.171 | -0.0436 | No | ||
| 146 | ACVR1B | 4335 22353 | 14601 | -0.180 | -0.0462 | No | ||
| 147 | PINK1 | 12752 15703 | 14650 | -0.185 | -0.0466 | No | ||
| 148 | ERN1 | 8137 | 14698 | -0.190 | -0.0470 | No | ||
| 149 | GSK3B | 22761 | 14988 | -0.227 | -0.0600 | No | ||
| 150 | WNK4 | 7644 | 15127 | -0.244 | -0.0646 | No | ||
| 151 | BRDT | 4327 | 15292 | -0.272 | -0.0703 | No | ||
| 152 | CDK10 | 3818 6157 10635 3894 | 15345 | -0.281 | -0.0699 | No | ||
| 153 | CSNK2A2 | 4567 8808 | 15415 | -0.291 | -0.0702 | No | ||
| 154 | LMTK2 | 16631 | 15467 | -0.303 | -0.0694 | No | ||
| 155 | SRPK2 | 5513 | 15688 | -0.354 | -0.0772 | No | ||
| 156 | BRSK2 | 190 | 15781 | -0.373 | -0.0779 | No | ||
| 157 | RPS6KA3 | 8490 | 15832 | -0.385 | -0.0761 | No | ||
| 158 | PIM2 | 5249 9564 9565 | 15952 | -0.417 | -0.0777 | No | ||
| 159 | UHMK1 | 4038 4957 | 16132 | -0.467 | -0.0819 | No | ||
| 160 | EIF2AK1 | 4869 | 16300 | -0.517 | -0.0849 | No | ||
| 161 | MAPK8 | 6459 | 16644 | -0.646 | -0.0960 | No | ||
| 162 | CDC42BPB | 20982 | 16817 | -0.728 | -0.0968 | No | ||
| 163 | PAK2 | 10310 5875 | 16924 | -0.783 | -0.0934 | No | ||
| 164 | ACVR1 | 4334 | 17118 | -0.886 | -0.0935 | No | ||
| 165 | MAPK1 | 1642 11167 | 17667 | -1.160 | -0.1097 | No | ||
| 166 | VRK2 | 947 20511 | 17714 | -1.192 | -0.0983 | No | ||
| 167 | BCKDK | 18061 1391 | 17800 | -1.254 | -0.0882 | No | ||
| 168 | PBK | 11959 | 17983 | -1.395 | -0.0818 | No | ||
| 169 | MAPK6 | 19062 | 18044 | -1.445 | -0.0682 | No | ||
| 170 | PRKAG1 | 22135 | 18092 | -1.496 | -0.0533 | No | ||
| 171 | STK19 | 23016 1569 1508 | 18130 | -1.543 | -0.0373 | No | ||
| 172 | PRKD3 | 13252 7956 | 18136 | -1.550 | -0.0194 | No | ||
| 173 | STK3 | 7084 | 18277 | -1.738 | -0.0067 | No | ||
| 174 | RIPK3 | 21817 | 18461 | -2.145 | 0.0084 | No |