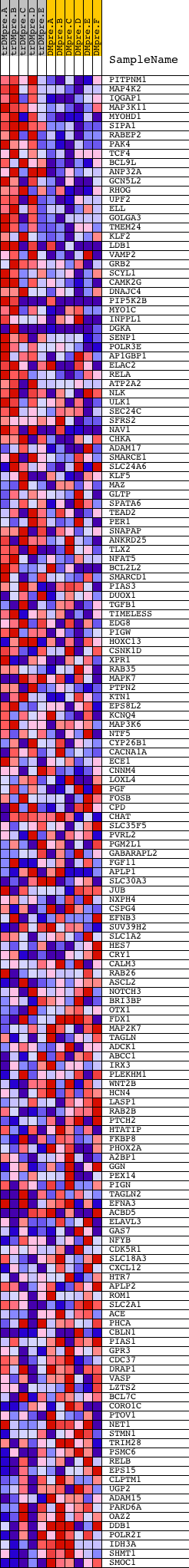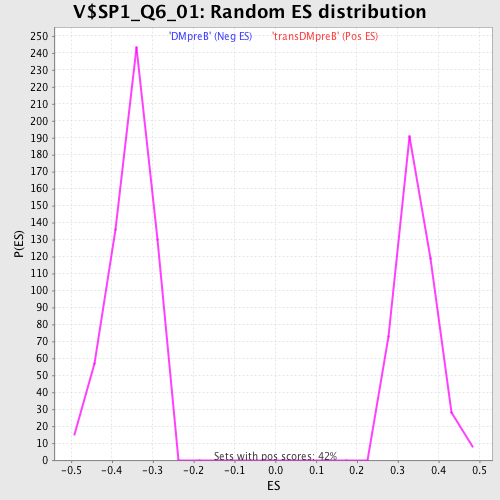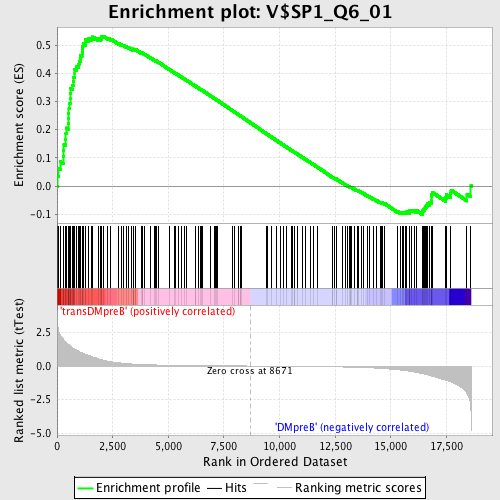
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | V$SP1_Q6_01 |
| Enrichment Score (ES) | 0.5320531 |
| Normalized Enrichment Score (NES) | 1.5479555 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.18467557 |
| FWER p-Value | 0.62 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PITPNM1 | 23770 | 20 | 3.033 | 0.0351 | Yes | ||
| 2 | MAP4K2 | 6457 | 72 | 2.603 | 0.0635 | Yes | ||
| 3 | IQGAP1 | 6619 | 136 | 2.288 | 0.0874 | Yes | ||
| 4 | MAP3K11 | 11163 | 275 | 2.001 | 0.1038 | Yes | ||
| 5 | MYOHD1 | 20724 | 284 | 1.966 | 0.1269 | Yes | ||
| 6 | SIPA1 | 9820 | 307 | 1.908 | 0.1485 | Yes | ||
| 7 | RABEP2 | 18082 | 373 | 1.801 | 0.1665 | Yes | ||
| 8 | PAK4 | 17909 | 396 | 1.777 | 0.1865 | Yes | ||
| 9 | TCF4 | 5642 | 417 | 1.735 | 0.2061 | Yes | ||
| 10 | BCL9L | 19475 | 495 | 1.633 | 0.2215 | Yes | ||
| 11 | ANP32A | 8592 4387 19415 | 514 | 1.618 | 0.2398 | Yes | ||
| 12 | GCN5L2 | 4763 | 521 | 1.601 | 0.2586 | Yes | ||
| 13 | RHOG | 17727 | 533 | 1.576 | 0.2769 | Yes | ||
| 14 | UPF2 | 11591 | 550 | 1.558 | 0.2946 | Yes | ||
| 15 | ELL | 18586 | 584 | 1.520 | 0.3110 | Yes | ||
| 16 | GOLGA3 | 6535 | 590 | 1.508 | 0.3287 | Yes | ||
| 17 | TMEM24 | 19150 | 600 | 1.490 | 0.3460 | Yes | ||
| 18 | KLF2 | 9229 | 686 | 1.384 | 0.3580 | Yes | ||
| 19 | LDB1 | 23654 | 722 | 1.333 | 0.3720 | Yes | ||
| 20 | VAMP2 | 20826 | 750 | 1.302 | 0.3861 | Yes | ||
| 21 | GRB2 | 20149 | 792 | 1.266 | 0.3990 | Yes | ||
| 22 | SCYL1 | 23986 | 795 | 1.264 | 0.4140 | Yes | ||
| 23 | CAMK2G | 21905 | 866 | 1.191 | 0.4244 | Yes | ||
| 24 | DNAJC4 | 23796 | 959 | 1.093 | 0.4325 | Yes | ||
| 25 | PIP5K2B | 20269 1254 | 988 | 1.061 | 0.4436 | Yes | ||
| 26 | MYO1C | 20777 | 1045 | 1.018 | 0.4528 | Yes | ||
| 27 | INPPL1 | 17733 3775 | 1055 | 1.014 | 0.4644 | Yes | ||
| 28 | DGKA | 3359 19589 | 1121 | 0.990 | 0.4727 | Yes | ||
| 29 | SENP1 | 10301 | 1136 | 0.983 | 0.4837 | Yes | ||
| 30 | POLR3E | 18100 | 1152 | 0.971 | 0.4945 | Yes | ||
| 31 | AP1GBP1 | 20725 | 1166 | 0.960 | 0.5052 | Yes | ||
| 32 | ELAC2 | 1267 20843 | 1266 | 0.884 | 0.5104 | Yes | ||
| 33 | RELA | 23783 | 1277 | 0.876 | 0.5203 | Yes | ||
| 34 | ATP2A2 | 4421 3481 | 1416 | 0.790 | 0.5223 | Yes | ||
| 35 | NLK | 5179 5178 | 1529 | 0.723 | 0.5249 | Yes | ||
| 36 | ULK1 | 16436 | 1603 | 0.681 | 0.5291 | Yes | ||
| 37 | SEC24C | 22086 | 1839 | 0.542 | 0.5228 | Yes | ||
| 38 | SFRS2 | 9807 20136 | 1971 | 0.471 | 0.5213 | Yes | ||
| 39 | NAV1 | 3978 5681 | 1980 | 0.468 | 0.5265 | Yes | ||
| 40 | CHKA | 23759 | 1991 | 0.465 | 0.5315 | Yes | ||
| 41 | ADAM17 | 4343 | 2076 | 0.429 | 0.5321 | Yes | ||
| 42 | SMARCE1 | 1218 20253 | 2283 | 0.359 | 0.5252 | No | ||
| 43 | SLC24A6 | 16721 | 2416 | 0.318 | 0.5218 | No | ||
| 44 | KLF5 | 4456 | 2761 | 0.240 | 0.5060 | No | ||
| 45 | MAZ | 1327 17623 | 2901 | 0.216 | 0.5011 | No | ||
| 46 | GLTP | 16419 | 2963 | 0.206 | 0.5002 | No | ||
| 47 | SPATA6 | 16134 | 3102 | 0.186 | 0.4950 | No | ||
| 48 | TEAD2 | 18252 | 3224 | 0.168 | 0.4904 | No | ||
| 49 | PER1 | 20827 | 3332 | 0.155 | 0.4865 | No | ||
| 50 | SNAPAP | 15270 | 3411 | 0.147 | 0.4840 | No | ||
| 51 | ANKRD25 | 6161 | 3430 | 0.144 | 0.4848 | No | ||
| 52 | TLX2 | 5773 1165 | 3433 | 0.144 | 0.4864 | No | ||
| 53 | NFAT5 | 3921 7037 12036 | 3519 | 0.137 | 0.4834 | No | ||
| 54 | BCL2L2 | 8653 4441 | 3543 | 0.135 | 0.4838 | No | ||
| 55 | SMARCD1 | 2241 22360 2250 | 3772 | 0.116 | 0.4728 | No | ||
| 56 | PIAS3 | 15491 672 1906 | 3825 | 0.113 | 0.4713 | No | ||
| 57 | DUOX1 | 14879 | 3831 | 0.112 | 0.4724 | No | ||
| 58 | TGFB1 | 18332 | 3946 | 0.105 | 0.4675 | No | ||
| 59 | TIMELESS | 10177 | 4208 | 0.089 | 0.4544 | No | ||
| 60 | EDG8 | 19212 | 4387 | 0.081 | 0.4457 | No | ||
| 61 | PIGW | 20312 | 4430 | 0.079 | 0.4444 | No | ||
| 62 | HOXC13 | 22344 | 4449 | 0.078 | 0.4443 | No | ||
| 63 | CSNK1D | 4181 8352 1387 | 4572 | 0.073 | 0.4386 | No | ||
| 64 | XPR1 | 5383 | 5038 | 0.058 | 0.4141 | No | ||
| 65 | RAB35 | 13386 | 5275 | 0.051 | 0.4019 | No | ||
| 66 | MAPK7 | 1381 20414 | 5307 | 0.050 | 0.4008 | No | ||
| 67 | PTPN2 | 23414 | 5435 | 0.047 | 0.3945 | No | ||
| 68 | KTN1 | 2865 22034 2586 | 5605 | 0.044 | 0.3859 | No | ||
| 69 | EPS8L2 | 18012 | 5719 | 0.041 | 0.3803 | No | ||
| 70 | KCNQ4 | 12247 2533 | 5808 | 0.040 | 0.3760 | No | ||
| 71 | MAP3K6 | 16054 | 6199 | 0.033 | 0.3552 | No | ||
| 72 | NTF5 | 18248 | 6200 | 0.033 | 0.3556 | No | ||
| 73 | CYP26B1 | 17093 | 6349 | 0.030 | 0.3480 | No | ||
| 74 | CACNA1A | 4461 | 6455 | 0.028 | 0.3426 | No | ||
| 75 | ECE1 | 2427 10516 | 6488 | 0.028 | 0.3412 | No | ||
| 76 | CNNM4 | 14273 | 6544 | 0.027 | 0.3385 | No | ||
| 77 | LOXL4 | 23672 | 6916 | 0.021 | 0.3187 | No | ||
| 78 | PGF | 21020 | 7086 | 0.019 | 0.3098 | No | ||
| 79 | FOSB | 17945 | 7115 | 0.019 | 0.3085 | No | ||
| 80 | CPD | 20344 4556 | 7162 | 0.018 | 0.3062 | No | ||
| 81 | CHAT | 21882 | 7211 | 0.017 | 0.3038 | No | ||
| 82 | SLC35F5 | 13171 | 7900 | 0.009 | 0.2666 | No | ||
| 83 | PVRL2 | 9671 5336 | 7985 | 0.008 | 0.2622 | No | ||
| 84 | PGM2L1 | 7712 | 8142 | 0.006 | 0.2538 | No | ||
| 85 | GABARAPL2 | 8211 13552 | 8166 | 0.006 | 0.2526 | No | ||
| 86 | FGF11 | 20383 | 8220 | 0.005 | 0.2498 | No | ||
| 87 | APLP1 | 17890 | 8228 | 0.005 | 0.2495 | No | ||
| 88 | SLC30A3 | 5995 | 8291 | 0.004 | 0.2462 | No | ||
| 89 | JUB | 21834 3339 | 9413 | -0.009 | 0.1855 | No | ||
| 90 | NXPH4 | 19602 | 9438 | -0.010 | 0.1844 | No | ||
| 91 | CSPG4 | 19435 15262 | 9646 | -0.012 | 0.1733 | No | ||
| 92 | EFNB3 | 20391 | 9854 | -0.015 | 0.1622 | No | ||
| 93 | SUV39H2 | 7232 12263 | 10059 | -0.018 | 0.1514 | No | ||
| 94 | SLC1A2 | 5453 | 10174 | -0.019 | 0.1455 | No | ||
| 95 | HES7 | 20828 | 10319 | -0.021 | 0.1379 | No | ||
| 96 | CRY1 | 19662 | 10517 | -0.024 | 0.1275 | No | ||
| 97 | CALM3 | 8682 | 10585 | -0.025 | 0.1242 | No | ||
| 98 | RAB26 | 11609 | 10665 | -0.026 | 0.1202 | No | ||
| 99 | ASCL2 | 5079 | 10681 | -0.026 | 0.1197 | No | ||
| 100 | NOTCH3 | 23031 | 10826 | -0.028 | 0.1123 | No | ||
| 101 | BRI3BP | 16696 | 11011 | -0.031 | 0.1027 | No | ||
| 102 | OTX1 | 20517 | 11153 | -0.033 | 0.0954 | No | ||
| 103 | FDX1 | 19117 | 11370 | -0.037 | 0.0841 | No | ||
| 104 | MAP2K7 | 6453 | 11507 | -0.039 | 0.0772 | No | ||
| 105 | TAGLN | 19136 | 11725 | -0.043 | 0.0660 | No | ||
| 106 | ADCK1 | 21191 | 12372 | -0.058 | 0.0317 | No | ||
| 107 | ABCC1 | 1668 5087 | 12385 | -0.058 | 0.0317 | No | ||
| 108 | IRX3 | 18793 | 12480 | -0.061 | 0.0274 | No | ||
| 109 | PLEKHM1 | 20196 1449 | 12535 | -0.063 | 0.0252 | No | ||
| 110 | WNT2B | 15214 1838 | 12537 | -0.063 | 0.0259 | No | ||
| 111 | HCN4 | 9080 | 12825 | -0.072 | 0.0112 | No | ||
| 112 | LASP1 | 4986 4985 1248 | 12967 | -0.077 | 0.0045 | No | ||
| 113 | RAB2B | 21842 | 13034 | -0.079 | 0.0018 | No | ||
| 114 | PTCH2 | 9640 | 13134 | -0.083 | -0.0025 | No | ||
| 115 | HTATIP | 3690 | 13190 | -0.085 | -0.0045 | No | ||
| 116 | FKBP8 | 18587 | 13201 | -0.085 | -0.0040 | No | ||
| 117 | PHOX2A | 18168 | 13251 | -0.087 | -0.0056 | No | ||
| 118 | A2BP1 | 11212 1652 1689 | 13356 | -0.092 | -0.0102 | No | ||
| 119 | GGN | 2472 1171 18312 | 13481 | -0.098 | -0.0157 | No | ||
| 120 | PEX14 | 15668 | 13499 | -0.099 | -0.0155 | No | ||
| 121 | PIGN | 13865 | 13530 | -0.100 | -0.0159 | No | ||
| 122 | TAGLN2 | 14043 | 13667 | -0.108 | -0.0220 | No | ||
| 123 | EFNA3 | 15278 | 13781 | -0.114 | -0.0267 | No | ||
| 124 | ACBD5 | 15105 2751 | 13941 | -0.124 | -0.0339 | No | ||
| 125 | ELAVL3 | 9136 | 14054 | -0.131 | -0.0384 | No | ||
| 126 | GAS7 | 20836 1299 | 14223 | -0.144 | -0.0458 | No | ||
| 127 | NFYB | 5173 | 14361 | -0.155 | -0.0513 | No | ||
| 128 | CDK5R1 | 20745 | 14516 | -0.171 | -0.0576 | No | ||
| 129 | SLC18A3 | 21883 | 14583 | -0.178 | -0.0591 | No | ||
| 130 | CXCL12 | 9792 1182 1031 | 14611 | -0.181 | -0.0584 | No | ||
| 131 | HTR7 | 23690 | 14729 | -0.194 | -0.0624 | No | ||
| 132 | APLP2 | 19190 | 15312 | -0.276 | -0.0907 | No | ||
| 133 | ROM1 | 23751 | 15429 | -0.294 | -0.0934 | No | ||
| 134 | SLC2A1 | 16107 | 15504 | -0.309 | -0.0938 | No | ||
| 135 | ACE | 1419 4313 | 15573 | -0.326 | -0.0935 | No | ||
| 136 | PHCA | 12301 7263 | 15640 | -0.340 | -0.0931 | No | ||
| 137 | CBLN1 | 18800 | 15687 | -0.354 | -0.0913 | No | ||
| 138 | PIAS1 | 7126 | 15828 | -0.383 | -0.0943 | No | ||
| 139 | GPR3 | 15729 | 15836 | -0.386 | -0.0901 | No | ||
| 140 | CDC37 | 19214 | 15852 | -0.390 | -0.0863 | No | ||
| 141 | DRAP1 | 23979 | 15948 | -0.416 | -0.0864 | No | ||
| 142 | VASP | 5847 | 16051 | -0.445 | -0.0867 | No | ||
| 143 | LZTS2 | 10362 | 16172 | -0.479 | -0.0874 | No | ||
| 144 | BCL7C | 4442 | 16419 | -0.559 | -0.0941 | No | ||
| 145 | CORO1C | 16425 | 16433 | -0.562 | -0.0881 | No | ||
| 146 | PTOV1 | 13522 | 16483 | -0.581 | -0.0838 | No | ||
| 147 | NET1 | 7100 | 16515 | -0.593 | -0.0784 | No | ||
| 148 | STMN1 | 9261 | 16546 | -0.609 | -0.0727 | No | ||
| 149 | TRIM28 | 18389 | 16604 | -0.634 | -0.0683 | No | ||
| 150 | PSMC6 | 7379 | 16655 | -0.652 | -0.0632 | No | ||
| 151 | RELB | 17942 | 16740 | -0.696 | -0.0594 | No | ||
| 152 | EPS15 | 4677 | 16815 | -0.728 | -0.0547 | No | ||
| 153 | CLPTM1 | 17940 | 16822 | -0.729 | -0.0463 | No | ||
| 154 | UGP2 | 20518 | 16824 | -0.730 | -0.0377 | No | ||
| 155 | ADAM15 | 15275 | 16836 | -0.739 | -0.0294 | No | ||
| 156 | PARD6A | 12142 | 16854 | -0.748 | -0.0214 | No | ||
| 157 | OAZ2 | 9499 | 17454 | -1.039 | -0.0415 | No | ||
| 158 | DDB1 | 23931 | 17493 | -1.060 | -0.0309 | No | ||
| 159 | POLR2I | 12839 | 17691 | -1.175 | -0.0275 | No | ||
| 160 | IDH3A | 19447 | 17703 | -1.181 | -0.0140 | No | ||
| 161 | SHMT1 | 5431 | 18423 | -2.013 | -0.0289 | No | ||
| 162 | SMOC1 | 21221 | 18602 | -3.291 | 0.0008 | No |