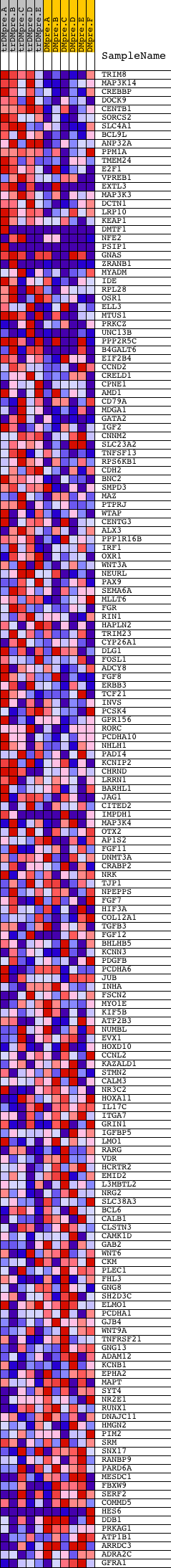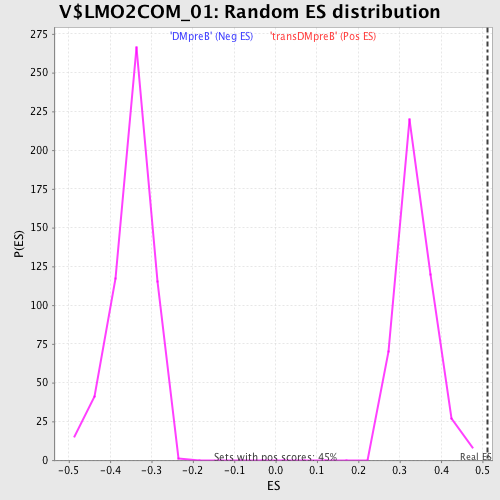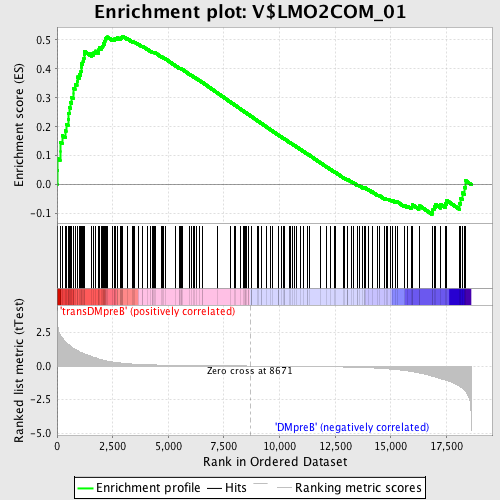
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | V$LMO2COM_01 |
| Enrichment Score (ES) | 0.5130813 |
| Normalized Enrichment Score (NES) | 1.5165662 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.16863576 |
| FWER p-Value | 0.763 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TRIM8 | 23824 | 5 | 3.715 | 0.0503 | Yes | ||
| 2 | MAP3K14 | 11998 | 27 | 2.958 | 0.0894 | Yes | ||
| 3 | CREBBP | 22682 8783 | 133 | 2.312 | 0.1152 | Yes | ||
| 4 | DOCK9 | 8369 | 135 | 2.291 | 0.1463 | Yes | ||
| 5 | CENTB1 | 20378 | 228 | 2.096 | 0.1698 | Yes | ||
| 6 | SORCS2 | 16553 | 382 | 1.794 | 0.1859 | Yes | ||
| 7 | SLC4A1 | 20204 | 402 | 1.763 | 0.2089 | Yes | ||
| 8 | BCL9L | 19475 | 495 | 1.633 | 0.2262 | Yes | ||
| 9 | ANP32A | 8592 4387 19415 | 514 | 1.618 | 0.2472 | Yes | ||
| 10 | PPM1A | 5279 9607 | 556 | 1.551 | 0.2661 | Yes | ||
| 11 | TMEM24 | 19150 | 600 | 1.490 | 0.2840 | Yes | ||
| 12 | E2F1 | 14384 | 657 | 1.419 | 0.3003 | Yes | ||
| 13 | VPREB1 | 5859 | 723 | 1.332 | 0.3149 | Yes | ||
| 14 | EXTL3 | 21785 21786 3088 | 724 | 1.330 | 0.3330 | Yes | ||
| 15 | MAP3K3 | 20626 | 808 | 1.254 | 0.3456 | Yes | ||
| 16 | DCTN1 | 1100 17392 | 895 | 1.166 | 0.3568 | Yes | ||
| 17 | LRP10 | 22018 | 906 | 1.156 | 0.3720 | Yes | ||
| 18 | KEAP1 | 19213 3080 3142 | 993 | 1.058 | 0.3817 | Yes | ||
| 19 | DMTF1 | 887 3662 16928 3530 | 1067 | 1.009 | 0.3915 | Yes | ||
| 20 | NFE2 | 9456 | 1087 | 1.001 | 0.4041 | Yes | ||
| 21 | PSIP1 | 4159 2507 8319 | 1095 | 0.999 | 0.4173 | Yes | ||
| 22 | GNAS | 9025 2963 2752 | 1162 | 0.961 | 0.4268 | Yes | ||
| 23 | ZRANB1 | 6884 11702 | 1201 | 0.927 | 0.4374 | Yes | ||
| 24 | MYADM | 11946 | 1215 | 0.920 | 0.4492 | Yes | ||
| 25 | IDE | 4893 | 1227 | 0.912 | 0.4610 | Yes | ||
| 26 | RPL28 | 5392 | 1555 | 0.704 | 0.4529 | Yes | ||
| 27 | OSR1 | 18971 | 1655 | 0.653 | 0.4564 | Yes | ||
| 28 | ELL3 | 14455 | 1713 | 0.620 | 0.4617 | Yes | ||
| 29 | MTUS1 | 4162 158 | 1859 | 0.533 | 0.4611 | Yes | ||
| 30 | PRKCZ | 5260 | 1877 | 0.520 | 0.4673 | Yes | ||
| 31 | UNC13B | 10255 | 1907 | 0.503 | 0.4725 | Yes | ||
| 32 | PPP2R5C | 6522 2147 | 1988 | 0.466 | 0.4745 | Yes | ||
| 33 | B4GALT6 | 7109 12115 | 2031 | 0.446 | 0.4783 | Yes | ||
| 34 | EIF2B4 | 16574 | 2064 | 0.435 | 0.4825 | Yes | ||
| 35 | CCND2 | 16987 | 2088 | 0.425 | 0.4871 | Yes | ||
| 36 | CRELD1 | 17329 | 2128 | 0.409 | 0.4905 | Yes | ||
| 37 | CPNE1 | 11186 | 2135 | 0.406 | 0.4957 | Yes | ||
| 38 | AMD1 | 8583 | 2155 | 0.398 | 0.5001 | Yes | ||
| 39 | CD79A | 18342 | 2161 | 0.395 | 0.5052 | Yes | ||
| 40 | MDGA1 | 13231 | 2201 | 0.384 | 0.5083 | Yes | ||
| 41 | GATA2 | 17370 | 2259 | 0.366 | 0.5102 | Yes | ||
| 42 | IGF2 | 17550 | 2506 | 0.291 | 0.5008 | Yes | ||
| 43 | CNNM2 | 8251 | 2573 | 0.276 | 0.5010 | Yes | ||
| 44 | SLC23A2 | 14421 | 2598 | 0.270 | 0.5034 | Yes | ||
| 45 | TNFSF13 | 12806 | 2618 | 0.265 | 0.5059 | Yes | ||
| 46 | RPS6KB1 | 7815 1207 13040 | 2699 | 0.250 | 0.5050 | Yes | ||
| 47 | CDH2 | 1963 8727 8726 4508 | 2701 | 0.250 | 0.5084 | Yes | ||
| 48 | BNC2 | 15850 | 2836 | 0.228 | 0.5042 | Yes | ||
| 49 | SMPD3 | 18757 | 2848 | 0.226 | 0.5067 | Yes | ||
| 50 | MAZ | 1327 17623 | 2901 | 0.216 | 0.5068 | Yes | ||
| 51 | PTPRJ | 9664 | 2913 | 0.213 | 0.5091 | Yes | ||
| 52 | WTAP | 7215 | 2917 | 0.212 | 0.5118 | Yes | ||
| 53 | CENTG3 | 3462 16905 3596 | 2947 | 0.208 | 0.5131 | Yes | ||
| 54 | ALX3 | 15456 | 3163 | 0.177 | 0.5038 | No | ||
| 55 | PPP1R16B | 6014 | 3381 | 0.149 | 0.4941 | No | ||
| 56 | IRF1 | 1336 1258 1433 9182 | 3435 | 0.144 | 0.4932 | No | ||
| 57 | OXR1 | 9300 2271 | 3497 | 0.138 | 0.4917 | No | ||
| 58 | WNT3A | 20431 | 3647 | 0.126 | 0.4854 | No | ||
| 59 | NEURL | 5164 | 3823 | 0.113 | 0.4774 | No | ||
| 60 | PAX9 | 21266 | 3859 | 0.110 | 0.4770 | No | ||
| 61 | SEMA6A | 23439 9802 | 4042 | 0.100 | 0.4685 | No | ||
| 62 | MLLT6 | 20678 | 4195 | 0.090 | 0.4615 | No | ||
| 63 | FGR | 4723 | 4299 | 0.084 | 0.4571 | No | ||
| 64 | RIN1 | 10349 | 4300 | 0.084 | 0.4582 | No | ||
| 65 | HAPLN2 | 15296 | 4351 | 0.082 | 0.4566 | No | ||
| 66 | TRIM23 | 8165 3230 | 4389 | 0.081 | 0.4557 | No | ||
| 67 | CYP26A1 | 23871 3695 | 4425 | 0.079 | 0.4549 | No | ||
| 68 | DLG1 | 22793 1672 | 4679 | 0.069 | 0.4421 | No | ||
| 69 | FOSL1 | 23779 | 4717 | 0.068 | 0.4410 | No | ||
| 70 | ADCY8 | 22281 | 4797 | 0.065 | 0.4376 | No | ||
| 71 | FGF8 | 23656 | 4879 | 0.063 | 0.4341 | No | ||
| 72 | ERBB3 | 19593 | 5331 | 0.050 | 0.4103 | No | ||
| 73 | TCF21 | 3437 | 5487 | 0.046 | 0.4025 | No | ||
| 74 | INVS | 2397 16211 | 5508 | 0.046 | 0.4021 | No | ||
| 75 | PCSK4 | 19696 3296 3394 18705 | 5555 | 0.045 | 0.4002 | No | ||
| 76 | GPR156 | 10753 22763 | 5577 | 0.044 | 0.3997 | No | ||
| 77 | RORC | 9732 | 5591 | 0.044 | 0.3995 | No | ||
| 78 | PCDHA10 | 8792 | 5650 | 0.043 | 0.3970 | No | ||
| 79 | NHLH1 | 13754 | 5941 | 0.037 | 0.3818 | No | ||
| 80 | PADI4 | 2424 9547 | 6026 | 0.035 | 0.3777 | No | ||
| 81 | KCNIP2 | 23655 3725 | 6122 | 0.034 | 0.3730 | No | ||
| 82 | CHRND | 3931 4084 | 6182 | 0.033 | 0.3702 | No | ||
| 83 | LRRN1 | 17343 | 6282 | 0.031 | 0.3653 | No | ||
| 84 | BARHL1 | 14634 | 6379 | 0.030 | 0.3605 | No | ||
| 85 | JAG1 | 14415 | 6538 | 0.027 | 0.3523 | No | ||
| 86 | CITED2 | 5118 14477 | 6554 | 0.027 | 0.3519 | No | ||
| 87 | IMPDH1 | 17197 1131 | 7187 | 0.018 | 0.3178 | No | ||
| 88 | MAP3K4 | 23126 | 7801 | 0.010 | 0.2847 | No | ||
| 89 | OTX2 | 9519 | 7989 | 0.008 | 0.2747 | No | ||
| 90 | AP1S2 | 8423 4228 | 8028 | 0.007 | 0.2727 | No | ||
| 91 | FGF11 | 20383 | 8220 | 0.005 | 0.2624 | No | ||
| 92 | DNMT3A | 2167 21330 | 8385 | 0.003 | 0.2536 | No | ||
| 93 | CRABP2 | 15554 | 8426 | 0.003 | 0.2515 | No | ||
| 94 | NRK | 6559 | 8480 | 0.002 | 0.2486 | No | ||
| 95 | TJP1 | 17803 | 8481 | 0.002 | 0.2487 | No | ||
| 96 | NPEPPS | 20273 | 8508 | 0.002 | 0.2473 | No | ||
| 97 | FGF7 | 8965 | 8584 | 0.001 | 0.2432 | No | ||
| 98 | HIF3A | 11989 3784 2303 | 8606 | 0.001 | 0.2421 | No | ||
| 99 | COL12A1 | 3143 19054 | 8716 | -0.001 | 0.2362 | No | ||
| 100 | TGFB3 | 10161 | 8719 | -0.001 | 0.2361 | No | ||
| 101 | FGF12 | 1723 22621 | 8735 | -0.001 | 0.2353 | No | ||
| 102 | BHLHB5 | 15629 | 8991 | -0.004 | 0.2215 | No | ||
| 103 | KCNN3 | 15538 1913 | 9031 | -0.004 | 0.2194 | No | ||
| 104 | PDGFB | 22199 2311 | 9172 | -0.006 | 0.2119 | No | ||
| 105 | PCDHA6 | 2038 8788 | 9177 | -0.006 | 0.2118 | No | ||
| 106 | JUB | 21834 3339 | 9413 | -0.009 | 0.1992 | No | ||
| 107 | INHA | 14214 | 9587 | -0.011 | 0.1900 | No | ||
| 108 | FSCN2 | 20569 | 9675 | -0.013 | 0.1854 | No | ||
| 109 | MYO1E | 7742 | 9957 | -0.016 | 0.1704 | No | ||
| 110 | KIF5B | 4955 2031 | 9963 | -0.016 | 0.1704 | No | ||
| 111 | ATP2B3 | 24308 11557 | 9968 | -0.016 | 0.1704 | No | ||
| 112 | NUMBL | 1663 9496 | 10090 | -0.018 | 0.1640 | No | ||
| 113 | EVX1 | 17442 | 10160 | -0.019 | 0.1606 | No | ||
| 114 | HOXD10 | 14983 | 10206 | -0.020 | 0.1584 | No | ||
| 115 | CCNL2 | 15957 | 10239 | -0.020 | 0.1569 | No | ||
| 116 | KAZALD1 | 4206 | 10464 | -0.023 | 0.1451 | No | ||
| 117 | STMN2 | 15638 | 10503 | -0.024 | 0.1434 | No | ||
| 118 | CALM3 | 8682 | 10585 | -0.025 | 0.1393 | No | ||
| 119 | NR3C2 | 18562 13798 | 10682 | -0.026 | 0.1345 | No | ||
| 120 | HOXA11 | 17146 | 10778 | -0.028 | 0.1297 | No | ||
| 121 | IL17C | 18438 | 10951 | -0.030 | 0.1208 | No | ||
| 122 | ITGA7 | 19835 | 11089 | -0.032 | 0.1138 | No | ||
| 123 | GRIN1 | 9041 4804 | 11253 | -0.035 | 0.1054 | No | ||
| 124 | IGFBP5 | 4899 | 11262 | -0.035 | 0.1055 | No | ||
| 125 | LMO1 | 17685 | 11323 | -0.036 | 0.1027 | No | ||
| 126 | RARG | 22113 | 11828 | -0.045 | 0.0760 | No | ||
| 127 | VDR | 5852 10285 | 12129 | -0.052 | 0.0604 | No | ||
| 128 | HCRTR2 | 6907 | 12302 | -0.056 | 0.0519 | No | ||
| 129 | EMID2 | 16336 | 12468 | -0.061 | 0.0437 | No | ||
| 130 | L3MBTL2 | 22415 | 12491 | -0.061 | 0.0434 | No | ||
| 131 | NRG2 | 4274 | 12885 | -0.074 | 0.0231 | No | ||
| 132 | SLC38A3 | 13319 | 12937 | -0.076 | 0.0213 | No | ||
| 133 | BCL6 | 22624 | 13041 | -0.079 | 0.0168 | No | ||
| 134 | CALB1 | 4469 | 13044 | -0.080 | 0.0178 | No | ||
| 135 | CLSTN3 | 17006 | 13070 | -0.080 | 0.0175 | No | ||
| 136 | CAMK1D | 5977 2948 | 13212 | -0.086 | 0.0111 | No | ||
| 137 | GAB2 | 1821 18184 2025 | 13330 | -0.091 | 0.0060 | No | ||
| 138 | WNT6 | 5881 | 13500 | -0.099 | -0.0019 | No | ||
| 139 | CKM | 18362 | 13520 | -0.100 | -0.0015 | No | ||
| 140 | PLEC1 | 2273 2198 2205 2178 2230 22247 2213 2231 2295 2172 5263 2266 | 13573 | -0.103 | -0.0029 | No | ||
| 141 | FHL3 | 16092 2457 | 13741 | -0.112 | -0.0105 | No | ||
| 142 | GNG8 | 18378 | 13802 | -0.115 | -0.0122 | No | ||
| 143 | SH2D3C | 2864 | 13815 | -0.116 | -0.0112 | No | ||
| 144 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 13839 | -0.118 | -0.0109 | No | ||
| 145 | PCDHA1 | 8571 4368 | 13985 | -0.127 | -0.0170 | No | ||
| 146 | GJB4 | 4782 | 14162 | -0.138 | -0.0247 | No | ||
| 147 | WNT9A | 5709 | 14415 | -0.162 | -0.0361 | No | ||
| 148 | TNFRSF21 | 8244 | 14480 | -0.168 | -0.0373 | No | ||
| 149 | GNG13 | 23338 | 14728 | -0.194 | -0.0481 | No | ||
| 150 | ADAM12 | 2207 8545 | 14818 | -0.205 | -0.0501 | No | ||
| 151 | KCNB1 | 14334 | 14865 | -0.211 | -0.0497 | No | ||
| 152 | EPHA2 | 16006 | 14984 | -0.226 | -0.0531 | No | ||
| 153 | MAPT | 9420 1347 | 15056 | -0.234 | -0.0537 | No | ||
| 154 | SYT4 | 5566 | 15212 | -0.257 | -0.0586 | No | ||
| 155 | NR2E1 | 19780 | 15278 | -0.269 | -0.0585 | No | ||
| 156 | RUNX1 | 4481 | 15616 | -0.335 | -0.0722 | No | ||
| 157 | DNAJC11 | 2486 6068 | 15742 | -0.364 | -0.0740 | No | ||
| 158 | HMGN2 | 9095 | 15925 | -0.409 | -0.0783 | No | ||
| 159 | PIM2 | 5249 9564 9565 | 15952 | -0.417 | -0.0741 | No | ||
| 160 | SRM | 9890 2525 | 15969 | -0.422 | -0.0692 | No | ||
| 161 | SNX17 | 3495 16885 | 16267 | -0.504 | -0.0784 | No | ||
| 162 | RANBP9 | 21480 | 16304 | -0.518 | -0.0733 | No | ||
| 163 | PARD6A | 12142 | 16854 | -0.748 | -0.0929 | No | ||
| 164 | MESDC1 | 17771 | 16890 | -0.765 | -0.0844 | No | ||
| 165 | FBXW9 | 18540 | 16948 | -0.797 | -0.0766 | No | ||
| 166 | SERF2 | 11714 | 17008 | -0.832 | -0.0685 | No | ||
| 167 | COMMD5 | 7289 | 17239 | -0.944 | -0.0681 | No | ||
| 168 | HES6 | 13882 4058 | 17445 | -1.035 | -0.0652 | No | ||
| 169 | DDB1 | 23931 | 17493 | -1.060 | -0.0533 | No | ||
| 170 | PRKAG1 | 22135 | 18092 | -1.496 | -0.0654 | No | ||
| 171 | ATP1B1 | 4420 | 18129 | -1.539 | -0.0464 | No | ||
| 172 | ARRDC3 | 4191 | 18215 | -1.643 | -0.0286 | No | ||
| 173 | ADRA2C | 16869 | 18316 | -1.802 | -0.0095 | No | ||
| 174 | GFRA1 | 9015 | 18358 | -1.890 | 0.0140 | No |