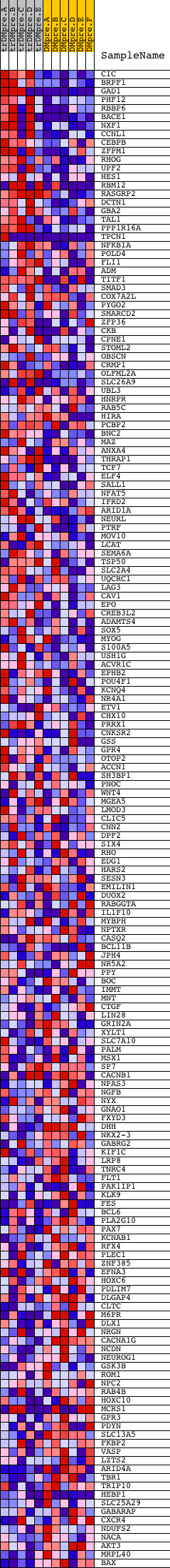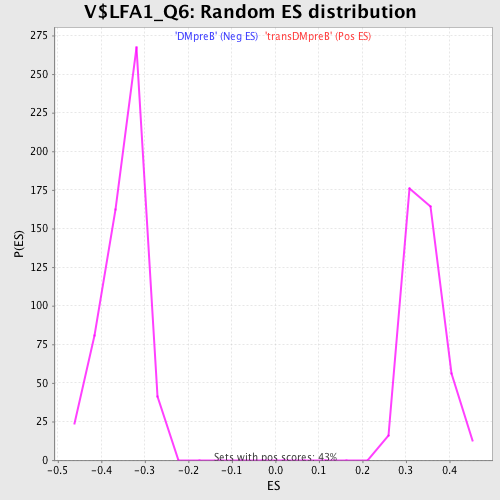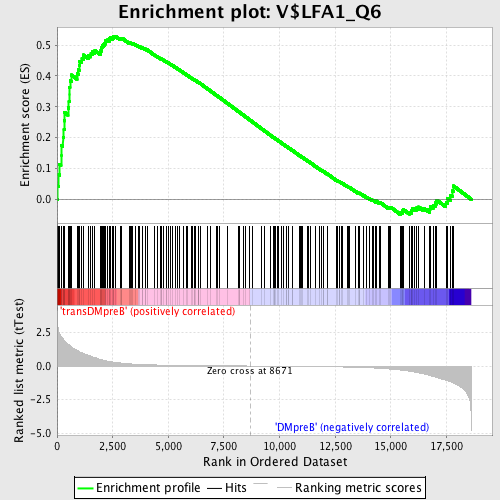
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | V$LFA1_Q6 |
| Enrichment Score (ES) | 0.52895385 |
| Normalized Enrichment Score (NES) | 1.5484345 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.20230244 |
| FWER p-Value | 0.613 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CIC | 18338 | 22 | 3.010 | 0.0435 | Yes | ||
| 2 | BRPF1 | 17332 | 71 | 2.621 | 0.0799 | Yes | ||
| 3 | GAD1 | 14993 2690 | 104 | 2.429 | 0.1143 | Yes | ||
| 4 | PHF12 | 11192 6483 | 175 | 2.205 | 0.1432 | Yes | ||
| 5 | RBBP6 | 5365 | 180 | 2.196 | 0.1757 | Yes | ||
| 6 | BACE1 | 6214 | 264 | 2.030 | 0.2013 | Yes | ||
| 7 | NXF1 | 11986 | 308 | 1.907 | 0.2274 | Yes | ||
| 8 | CCNL1 | 15325 | 323 | 1.872 | 0.2544 | Yes | ||
| 9 | CEBPB | 8733 | 335 | 1.856 | 0.2814 | Yes | ||
| 10 | ZFPM1 | 18439 | 490 | 1.637 | 0.2974 | Yes | ||
| 11 | RHOG | 17727 | 533 | 1.576 | 0.3185 | Yes | ||
| 12 | UPF2 | 11591 | 550 | 1.558 | 0.3408 | Yes | ||
| 13 | HES1 | 22798 | 551 | 1.558 | 0.3640 | Yes | ||
| 14 | RBM12 | 7980 2710 13280 | 580 | 1.524 | 0.3851 | Yes | ||
| 15 | RASGRP2 | 9693 | 641 | 1.433 | 0.4032 | Yes | ||
| 16 | DCTN1 | 1100 17392 | 895 | 1.166 | 0.4068 | Yes | ||
| 17 | GBA2 | 15898 | 957 | 1.094 | 0.4198 | Yes | ||
| 18 | TAL1 | 16129 | 992 | 1.059 | 0.4336 | Yes | ||
| 19 | PPP1R16A | 22441 | 1011 | 1.038 | 0.4481 | Yes | ||
| 20 | TPCN1 | 16398 6383 10915 | 1117 | 0.991 | 0.4571 | Yes | ||
| 21 | NFKBIA | 21065 | 1178 | 0.947 | 0.4680 | Yes | ||
| 22 | POLD4 | 12822 | 1404 | 0.798 | 0.4676 | Yes | ||
| 23 | FLI1 | 4729 | 1522 | 0.726 | 0.4721 | Yes | ||
| 24 | ADM | 18125 | 1588 | 0.688 | 0.4788 | Yes | ||
| 25 | TITF1 | 10180 21062 | 1699 | 0.629 | 0.4821 | Yes | ||
| 26 | SMAD3 | 19084 | 1948 | 0.484 | 0.4759 | Yes | ||
| 27 | COX7A2L | 9819 | 1965 | 0.474 | 0.4821 | Yes | ||
| 28 | PYGO2 | 12748 | 1997 | 0.462 | 0.4873 | Yes | ||
| 29 | SMARCD2 | 13515 | 2007 | 0.455 | 0.4935 | Yes | ||
| 30 | ZFP36 | 84 | 2050 | 0.439 | 0.4978 | Yes | ||
| 31 | CKB | 4523 | 2083 | 0.426 | 0.5024 | Yes | ||
| 32 | CPNE1 | 11186 | 2135 | 0.406 | 0.5056 | Yes | ||
| 33 | STOML2 | 15902 | 2180 | 0.388 | 0.5090 | Yes | ||
| 34 | OBSCN | 20434 | 2185 | 0.386 | 0.5145 | Yes | ||
| 35 | CRMP1 | 16860 | 2249 | 0.369 | 0.5166 | Yes | ||
| 36 | OLFML2A | 6304 | 2339 | 0.342 | 0.5169 | Yes | ||
| 37 | SLC26A9 | 11560 980 4067 | 2360 | 0.333 | 0.5207 | Yes | ||
| 38 | UBL3 | 16284 | 2395 | 0.324 | 0.5237 | Yes | ||
| 39 | HNRPR | 13196 7909 2500 | 2499 | 0.294 | 0.5225 | Yes | ||
| 40 | RAB5C | 20226 | 2527 | 0.286 | 0.5253 | Yes | ||
| 41 | HIRA | 4852 9090 | 2541 | 0.282 | 0.5288 | Yes | ||
| 42 | PCBP2 | 9533 | 2612 | 0.267 | 0.5290 | Yes | ||
| 43 | BNC2 | 15850 | 2836 | 0.228 | 0.5203 | No | ||
| 44 | MAZ | 1327 17623 | 2901 | 0.216 | 0.5200 | No | ||
| 45 | ANXA4 | 17083 | 2908 | 0.214 | 0.5228 | No | ||
| 46 | THRAP1 | 20309 | 3250 | 0.165 | 0.5068 | No | ||
| 47 | TCF7 | 1467 20466 | 3291 | 0.161 | 0.5070 | No | ||
| 48 | ELF4 | 24162 | 3341 | 0.153 | 0.5066 | No | ||
| 49 | SALL1 | 18796 12207 | 3408 | 0.147 | 0.5053 | No | ||
| 50 | NFAT5 | 3921 7037 12036 | 3519 | 0.137 | 0.5013 | No | ||
| 51 | IFRD2 | 19320 | 3655 | 0.125 | 0.4959 | No | ||
| 52 | ARID1A | 8214 2501 13554 | 3717 | 0.121 | 0.4943 | No | ||
| 53 | NEURL | 5164 | 3823 | 0.113 | 0.4903 | No | ||
| 54 | PTRF | 9667 | 3835 | 0.112 | 0.4914 | No | ||
| 55 | MOV10 | 9403 9402 5114 | 3960 | 0.105 | 0.4862 | No | ||
| 56 | LCAT | 18762 | 3968 | 0.104 | 0.4874 | No | ||
| 57 | SEMA6A | 23439 9802 | 4042 | 0.100 | 0.4849 | No | ||
| 58 | TSP50 | 19286 | 4395 | 0.080 | 0.4670 | No | ||
| 59 | SLC2A4 | 20380 | 4496 | 0.076 | 0.4627 | No | ||
| 60 | UQCRC1 | 10259 | 4631 | 0.071 | 0.4565 | No | ||
| 61 | LAG3 | 17000 | 4693 | 0.069 | 0.4542 | No | ||
| 62 | CAV1 | 8698 | 4694 | 0.069 | 0.4553 | No | ||
| 63 | EPO | 8911 | 4785 | 0.065 | 0.4514 | No | ||
| 64 | CREB3L2 | 17182 | 4924 | 0.061 | 0.4448 | No | ||
| 65 | ADAMTS4 | 10791 | 4925 | 0.061 | 0.4457 | No | ||
| 66 | SOX5 | 9850 1044 5480 16937 | 5007 | 0.058 | 0.4422 | No | ||
| 67 | MYOG | 14129 | 5100 | 0.056 | 0.4380 | No | ||
| 68 | S100A5 | 15529 | 5170 | 0.054 | 0.4351 | No | ||
| 69 | USH1G | 20153 | 5316 | 0.050 | 0.4280 | No | ||
| 70 | ACVR1C | 14585 | 5425 | 0.048 | 0.4228 | No | ||
| 71 | EPHB2 | 4675 2440 8910 | 5497 | 0.046 | 0.4196 | No | ||
| 72 | POU4F1 | 21727 | 5669 | 0.042 | 0.4110 | No | ||
| 73 | KCNQ4 | 12247 2533 | 5808 | 0.040 | 0.4041 | No | ||
| 74 | NR4A1 | 9099 | 5872 | 0.038 | 0.4013 | No | ||
| 75 | ETV1 | 4688 8925 8924 | 6060 | 0.035 | 0.3916 | No | ||
| 76 | CHX10 | 21208 | 6064 | 0.035 | 0.3920 | No | ||
| 77 | PRRX1 | 9595 5271 | 6195 | 0.033 | 0.3854 | No | ||
| 78 | CNKSR2 | 2553 24021 | 6196 | 0.033 | 0.3859 | No | ||
| 79 | GSS | 14380 9047 | 6204 | 0.032 | 0.3860 | No | ||
| 80 | GPR4 | 18364 | 6341 | 0.030 | 0.3791 | No | ||
| 81 | OTOP2 | 20600 | 6343 | 0.030 | 0.3795 | No | ||
| 82 | ACCN1 | 20327 1194 | 6365 | 0.030 | 0.3788 | No | ||
| 83 | SH3BP1 | 632 22431 | 6449 | 0.029 | 0.3747 | No | ||
| 84 | PNOC | 21783 | 6748 | 0.024 | 0.3589 | No | ||
| 85 | WNT4 | 16025 | 6894 | 0.022 | 0.3514 | No | ||
| 86 | MGEA5 | 8001 | 7150 | 0.018 | 0.3378 | No | ||
| 87 | LMOD3 | 17059 | 7209 | 0.017 | 0.3349 | No | ||
| 88 | CLIC5 | 23229 | 7317 | 0.016 | 0.3294 | No | ||
| 89 | CNN2 | 19953 3391 | 7656 | 0.012 | 0.3112 | No | ||
| 90 | DPF2 | 9719 | 8147 | 0.006 | 0.2847 | No | ||
| 91 | SIX4 | 5445 | 8186 | 0.006 | 0.2828 | No | ||
| 92 | RHO | 17314 | 8216 | 0.005 | 0.2813 | No | ||
| 93 | EDG1 | 15187 | 8396 | 0.003 | 0.2716 | No | ||
| 94 | HARS2 | 23592 14820 | 8487 | 0.002 | 0.2668 | No | ||
| 95 | SESN3 | 19561 | 8626 | 0.000 | 0.2593 | No | ||
| 96 | EMILIN1 | 16891 | 8786 | -0.001 | 0.2507 | No | ||
| 97 | DUOX2 | 14453 | 9174 | -0.006 | 0.2298 | No | ||
| 98 | RABGGTA | 2669 21820 | 9337 | -0.008 | 0.2211 | No | ||
| 99 | IL1F10 | 15097 | 9578 | -0.011 | 0.2083 | No | ||
| 100 | MYBPH | 14130 | 9737 | -0.014 | 0.1999 | No | ||
| 101 | NPTXR | 7854 22200 | 9748 | -0.014 | 0.1996 | No | ||
| 102 | CASQ2 | 15477 | 9785 | -0.014 | 0.1978 | No | ||
| 103 | BCL11B | 7190 | 9826 | -0.015 | 0.1959 | No | ||
| 104 | JPH4 | 21826 | 9921 | -0.016 | 0.1910 | No | ||
| 105 | NR5A2 | 4105 13817 | 9942 | -0.016 | 0.1902 | No | ||
| 106 | PPY | 20208 | 10104 | -0.018 | 0.1817 | No | ||
| 107 | BOC | 22587 | 10179 | -0.019 | 0.1780 | No | ||
| 108 | IMMT | 367 8029 | 10295 | -0.021 | 0.1720 | No | ||
| 109 | MNT | 20784 | 10331 | -0.021 | 0.1705 | No | ||
| 110 | CTGF | 20068 | 10415 | -0.022 | 0.1663 | No | ||
| 111 | LIN28 | 15723 | 10419 | -0.023 | 0.1665 | No | ||
| 112 | GRIN2A | 22668 | 10572 | -0.025 | 0.1586 | No | ||
| 113 | XYLT1 | 18113 | 10576 | -0.025 | 0.1588 | No | ||
| 114 | SLC7A10 | 18294 | 10916 | -0.030 | 0.1409 | No | ||
| 115 | PALM | 19960 3350 | 10943 | -0.030 | 0.1399 | No | ||
| 116 | MSX1 | 16547 | 11003 | -0.031 | 0.1372 | No | ||
| 117 | SP7 | 22110 | 11047 | -0.032 | 0.1353 | No | ||
| 118 | CACNB1 | 1443 1304 | 11273 | -0.035 | 0.1236 | No | ||
| 119 | NPAS3 | 2077 21273 | 11300 | -0.035 | 0.1227 | No | ||
| 120 | NGFB | 15476 | 11390 | -0.037 | 0.1185 | No | ||
| 121 | NYX | 24378 | 11623 | -0.041 | 0.1065 | No | ||
| 122 | GNAO1 | 4785 3829 | 11790 | -0.045 | 0.0982 | No | ||
| 123 | FXYD3 | 9370 | 11888 | -0.047 | 0.0936 | No | ||
| 124 | DHH | 4629 | 11960 | -0.048 | 0.0904 | No | ||
| 125 | NKX2-3 | 5177 23845 | 11963 | -0.048 | 0.0911 | No | ||
| 126 | GABRG2 | 1259 4748 8996 1331 | 12136 | -0.052 | 0.0825 | No | ||
| 127 | KIF1C | 4951 | 12174 | -0.053 | 0.0813 | No | ||
| 128 | LRP8 | 16149 2425 | 12550 | -0.063 | 0.0619 | No | ||
| 129 | TNRC4 | 15514 | 12615 | -0.065 | 0.0594 | No | ||
| 130 | FLT1 | 3483 16287 | 12672 | -0.067 | 0.0573 | No | ||
| 131 | PAK1IP1 | 21660 | 12694 | -0.068 | 0.0572 | No | ||
| 132 | KLK9 | 18267 | 12795 | -0.071 | 0.0528 | No | ||
| 133 | FES | 17779 3949 | 12837 | -0.072 | 0.0517 | No | ||
| 134 | BCL6 | 22624 | 13041 | -0.079 | 0.0419 | No | ||
| 135 | PLA2G10 | 22658 | 13105 | -0.082 | 0.0397 | No | ||
| 136 | PAX7 | 15697 | 13157 | -0.084 | 0.0381 | No | ||
| 137 | KCNAB1 | 1791 15577 | 13396 | -0.094 | 0.0266 | No | ||
| 138 | RFX4 | 7721 3410 | 13556 | -0.102 | 0.0195 | No | ||
| 139 | PLEC1 | 2273 2198 2205 2178 2230 22247 2213 2231 2295 2172 5263 2266 | 13573 | -0.103 | 0.0202 | No | ||
| 140 | ZNF385 | 22106 2297 | 13595 | -0.104 | 0.0206 | No | ||
| 141 | EFNA3 | 15278 | 13781 | -0.114 | 0.0122 | No | ||
| 142 | HOXC6 | 22339 2302 | 13907 | -0.121 | 0.0073 | No | ||
| 143 | PDLIM7 | 7434 | 14029 | -0.129 | 0.0026 | No | ||
| 144 | DLGAP4 | 2950 10461 | 14156 | -0.138 | -0.0021 | No | ||
| 145 | CLTC | 14610 | 14204 | -0.142 | -0.0026 | No | ||
| 146 | M6PR | 1053 5044 9328 | 14331 | -0.152 | -0.0072 | No | ||
| 147 | DLX1 | 14988 | 14336 | -0.152 | -0.0051 | No | ||
| 148 | NRGN | 19176 | 14474 | -0.167 | -0.0101 | No | ||
| 149 | CACNA1G | 1237 940 20292 | 14535 | -0.172 | -0.0107 | No | ||
| 150 | NCDN | 15756 2487 6471 | 14904 | -0.217 | -0.0275 | No | ||
| 151 | NEUROG1 | 21446 | 14958 | -0.222 | -0.0270 | No | ||
| 152 | GSK3B | 22761 | 14988 | -0.227 | -0.0252 | No | ||
| 153 | ROM1 | 23751 | 15429 | -0.294 | -0.0447 | No | ||
| 154 | NPC2 | 21023 | 15469 | -0.303 | -0.0423 | No | ||
| 155 | RAB4B | 5345 | 15519 | -0.313 | -0.0403 | No | ||
| 156 | HOXC10 | 22342 | 15539 | -0.316 | -0.0367 | No | ||
| 157 | MCRS1 | 6961 6960 6962 | 15568 | -0.324 | -0.0334 | No | ||
| 158 | GPR3 | 15729 | 15836 | -0.386 | -0.0421 | No | ||
| 159 | PDYN | 14429 | 15929 | -0.411 | -0.0410 | No | ||
| 160 | SLC13A5 | 133 | 15942 | -0.415 | -0.0355 | No | ||
| 161 | FKBP2 | 8972 | 15959 | -0.419 | -0.0301 | No | ||
| 162 | VASP | 5847 | 16051 | -0.445 | -0.0284 | No | ||
| 163 | LZTS2 | 10362 | 16172 | -0.479 | -0.0278 | No | ||
| 164 | ARID4A | 2080 6215 | 16253 | -0.500 | -0.0247 | No | ||
| 165 | TBR1 | 83 | 16493 | -0.584 | -0.0290 | No | ||
| 166 | TRIP10 | 23174 | 16748 | -0.697 | -0.0324 | No | ||
| 167 | HEBP1 | 16961 | 16762 | -0.705 | -0.0227 | No | ||
| 168 | SLC25A29 | 20985 | 16900 | -0.772 | -0.0186 | No | ||
| 169 | GABARAP | 20815 | 16994 | -0.823 | -0.0114 | No | ||
| 170 | CXCR4 | 13844 | 17067 | -0.864 | -0.0025 | No | ||
| 171 | NDUFS2 | 4089 13760 | 17480 | -1.054 | -0.0092 | No | ||
| 172 | NACA | 9444 5147 | 17545 | -1.084 | 0.0035 | No | ||
| 173 | AKT3 | 13739 982 | 17683 | -1.172 | 0.0134 | No | ||
| 174 | MRPL40 | 22641 | 17764 | -1.228 | 0.0274 | No | ||
| 175 | BAX | 17832 | 17815 | -1.264 | 0.0434 | No |