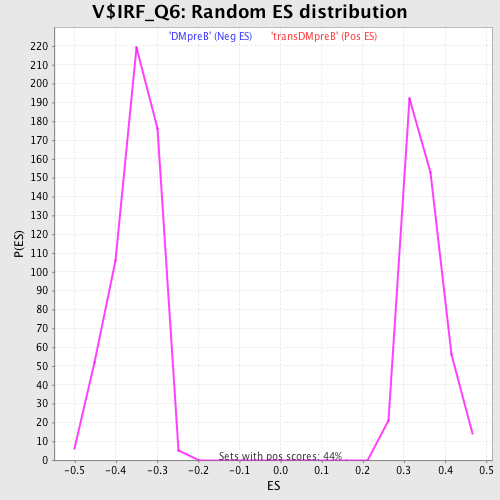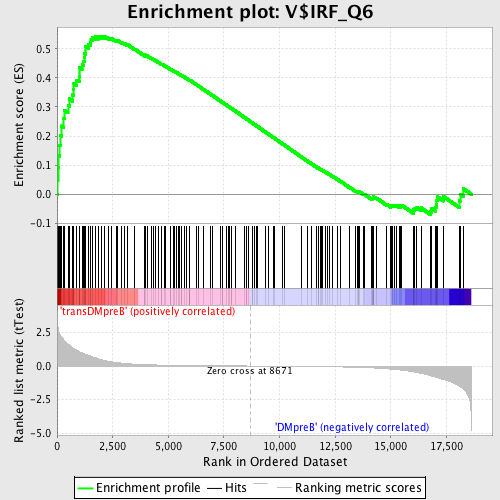
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | V$IRF_Q6 |
| Enrichment Score (ES) | 0.54331154 |
| Normalized Enrichment Score (NES) | 1.5725286 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.18860856 |
| FWER p-Value | 0.495 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BCL11A | 4691 | 19 | 3.045 | 0.0474 | Yes | ||
| 2 | LMO2 | 9281 | 40 | 2.855 | 0.0918 | Yes | ||
| 3 | ISGF3G | 2686 9184 | 59 | 2.692 | 0.1336 | Yes | ||
| 4 | LSP1 | 18003 | 120 | 2.366 | 0.1681 | Yes | ||
| 5 | RRBP1 | 8176 14411 | 156 | 2.233 | 0.2017 | Yes | ||
| 6 | ITGB7 | 22112 | 191 | 2.180 | 0.2346 | Yes | ||
| 7 | MAP3K11 | 11163 | 275 | 2.001 | 0.2619 | Yes | ||
| 8 | FCGR2B | 8959 | 336 | 1.855 | 0.2882 | Yes | ||
| 9 | ZFPM1 | 18439 | 490 | 1.637 | 0.3060 | Yes | ||
| 10 | UPF2 | 11591 | 550 | 1.558 | 0.3276 | Yes | ||
| 11 | CCND1 | 4487 4488 8707 17535 | 698 | 1.368 | 0.3414 | Yes | ||
| 12 | MS4A1 | 59 | 731 | 1.318 | 0.3606 | Yes | ||
| 13 | GSDMDC1 | 22453 12780 | 754 | 1.299 | 0.3801 | Yes | ||
| 14 | LGALS8 | 21544 | 886 | 1.179 | 0.3918 | Yes | ||
| 15 | PIP5K2B | 20269 1254 | 988 | 1.061 | 0.4032 | Yes | ||
| 16 | TOP1 | 5790 5789 10210 | 996 | 1.056 | 0.4196 | Yes | ||
| 17 | KYNU | 15016 | 997 | 1.056 | 0.4364 | Yes | ||
| 18 | DGKA | 3359 19589 | 1121 | 0.990 | 0.4455 | Yes | ||
| 19 | BBX | 7685 | 1204 | 0.926 | 0.4558 | Yes | ||
| 20 | PTMA | 9657 | 1221 | 0.916 | 0.4695 | Yes | ||
| 21 | TLK2 | 20628 | 1238 | 0.905 | 0.4831 | Yes | ||
| 22 | TLR7 | 24004 | 1274 | 0.878 | 0.4951 | Yes | ||
| 23 | SLC40A1 | 13964 | 1275 | 0.878 | 0.5091 | Yes | ||
| 24 | ADAR | 7116 | 1425 | 0.781 | 0.5135 | Yes | ||
| 25 | PTK2 | 22271 | 1498 | 0.741 | 0.5214 | Yes | ||
| 26 | FLI1 | 4729 | 1522 | 0.726 | 0.5317 | Yes | ||
| 27 | NLGN2 | 10117 | 1601 | 0.682 | 0.5383 | Yes | ||
| 28 | SELL | 9798 | 1721 | 0.617 | 0.5417 | Yes | ||
| 29 | IRF2 | 18621 | 1878 | 0.519 | 0.5415 | Yes | ||
| 30 | EHD1 | 23791 | 1983 | 0.467 | 0.5433 | Yes | ||
| 31 | IL18BP | 17730 | 2129 | 0.408 | 0.5420 | No | ||
| 32 | CASP7 | 23826 | 2325 | 0.347 | 0.5369 | No | ||
| 33 | ARHGEF6 | 13297 13104 | 2444 | 0.307 | 0.5354 | No | ||
| 34 | SLC39A7 | 23022 | 2652 | 0.259 | 0.5283 | No | ||
| 35 | DNASE1L3 | 21925 | 2720 | 0.247 | 0.5286 | No | ||
| 36 | PDGFRB | 23539 | 2909 | 0.214 | 0.5218 | No | ||
| 37 | USF1 | 5832 10260 10261 | 3036 | 0.195 | 0.5181 | No | ||
| 38 | RTN4 | 7569 1328 1343 1442 | 3144 | 0.181 | 0.5152 | No | ||
| 39 | PTPRO | 1093 17256 | 3491 | 0.139 | 0.4987 | No | ||
| 40 | STAT6 | 19854 9909 | 3938 | 0.106 | 0.4762 | No | ||
| 41 | TAPBP | 5625 | 3940 | 0.106 | 0.4779 | No | ||
| 42 | MOV10 | 9403 9402 5114 | 3960 | 0.105 | 0.4785 | No | ||
| 43 | ARF3 | 22137 | 4049 | 0.099 | 0.4753 | No | ||
| 44 | TBX1 | 10039 5630 | 4221 | 0.088 | 0.4675 | No | ||
| 45 | CDK6 | 16600 | 4318 | 0.083 | 0.4636 | No | ||
| 46 | SEMA3A | 16919 | 4428 | 0.079 | 0.4589 | No | ||
| 47 | BCOR | 12934 2561 | 4568 | 0.073 | 0.4526 | No | ||
| 48 | DCAMKL1 | 8838 4600 | 4700 | 0.068 | 0.4466 | No | ||
| 49 | TNFSF15 | 15863 | 4710 | 0.068 | 0.4472 | No | ||
| 50 | MIA2 | 21262 | 4843 | 0.064 | 0.4410 | No | ||
| 51 | LGI1 | 23867 | 4891 | 0.062 | 0.4395 | No | ||
| 52 | MYOG | 14129 | 5100 | 0.056 | 0.4291 | No | ||
| 53 | EREG | 4679 16797 | 5117 | 0.055 | 0.4291 | No | ||
| 54 | DAPP1 | 11159 6445 6446 | 5222 | 0.053 | 0.4243 | No | ||
| 55 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 5266 | 0.052 | 0.4228 | No | ||
| 56 | PITX2 | 15424 1878 | 5344 | 0.050 | 0.4194 | No | ||
| 57 | TCIRG1 | 3700 3752 23951 | 5464 | 0.047 | 0.4137 | No | ||
| 58 | IL22RA1 | 16037 | 5493 | 0.046 | 0.4130 | No | ||
| 59 | MLLT3 | 2359 15847 2413 | 5589 | 0.044 | 0.4085 | No | ||
| 60 | PLXNC1 | 7056 | 5609 | 0.043 | 0.4082 | No | ||
| 61 | B2M | 72 14882 | 5712 | 0.042 | 0.4033 | No | ||
| 62 | KCNQ4 | 12247 2533 | 5808 | 0.040 | 0.3988 | No | ||
| 63 | GATA6 | 96 | 5937 | 0.037 | 0.3925 | No | ||
| 64 | TNFSF13B | 6291 10792 | 5957 | 0.036 | 0.3920 | No | ||
| 65 | PCDH7 | 7029 3460 | 5963 | 0.036 | 0.3923 | No | ||
| 66 | HTR2C | 24230 | 6277 | 0.031 | 0.3759 | No | ||
| 67 | CASK | 24184 2618 2604 | 6366 | 0.030 | 0.3716 | No | ||
| 68 | PRDM16 | 12893 | 6569 | 0.027 | 0.3611 | No | ||
| 69 | CCNT2 | 3993 13079 | 6888 | 0.022 | 0.3442 | No | ||
| 70 | THBS1 | 5748 14910 | 7004 | 0.020 | 0.3383 | No | ||
| 71 | HOXB3 | 20685 | 7352 | 0.016 | 0.3197 | No | ||
| 72 | FMR1 | 24321 | 7412 | 0.015 | 0.3168 | No | ||
| 73 | RORB | 10356 | 7447 | 0.014 | 0.3152 | No | ||
| 74 | ECM2 | 300 | 7610 | 0.012 | 0.3066 | No | ||
| 75 | SIX1 | 9821 | 7711 | 0.011 | 0.3014 | No | ||
| 76 | MUSK | 5199 | 7752 | 0.010 | 0.2994 | No | ||
| 77 | SLC2A5 | 12138 | 7817 | 0.010 | 0.2960 | No | ||
| 78 | NCF1 | 16354 | 7826 | 0.010 | 0.2958 | No | ||
| 79 | IFNB1 | 15846 | 8029 | 0.007 | 0.2849 | No | ||
| 80 | EIF4A2 | 4660 1679 1645 | 8430 | 0.003 | 0.2633 | No | ||
| 81 | TWIST1 | 21291 | 8501 | 0.002 | 0.2596 | No | ||
| 82 | NPEPPS | 20273 | 8508 | 0.002 | 0.2593 | No | ||
| 83 | CDH3 | 3919 4509 | 8609 | 0.001 | 0.2539 | No | ||
| 84 | CHST1 | 13375 | 8773 | -0.001 | 0.2451 | No | ||
| 85 | FUT8 | 2163 21233 | 8800 | -0.002 | 0.2437 | No | ||
| 86 | DIO2 | 21014 2159 | 8884 | -0.002 | 0.2392 | No | ||
| 87 | HPCAL1 | 21307 2123 | 8958 | -0.003 | 0.2353 | No | ||
| 88 | BHLHB5 | 15629 | 8991 | -0.004 | 0.2337 | No | ||
| 89 | KLHL13 | 12536 | 9385 | -0.009 | 0.2125 | No | ||
| 90 | PURA | 9670 | 9498 | -0.010 | 0.2066 | No | ||
| 91 | GJC1 | 20256 | 9730 | -0.013 | 0.1943 | No | ||
| 92 | NEDD4 | 19384 3101 | 9763 | -0.014 | 0.1928 | No | ||
| 93 | CNTFR | 2515 15906 | 10148 | -0.019 | 0.1723 | No | ||
| 94 | TCF7L2 | 10048 5646 | 10216 | -0.020 | 0.1690 | No | ||
| 95 | MSX1 | 16547 | 11003 | -0.031 | 0.1269 | No | ||
| 96 | HIPK1 | 4851 | 11268 | -0.035 | 0.1132 | No | ||
| 97 | CBX4 | 20128 | 11439 | -0.038 | 0.1046 | No | ||
| 98 | PSMA5 | 6464 | 11660 | -0.042 | 0.0934 | No | ||
| 99 | ADORA3 | 4352 | 11727 | -0.043 | 0.0905 | No | ||
| 100 | ARHGAP5 | 4412 8625 | 11744 | -0.044 | 0.0903 | No | ||
| 101 | RARG | 22113 | 11828 | -0.045 | 0.0865 | No | ||
| 102 | DLL1 | 23119 | 11894 | -0.047 | 0.0837 | No | ||
| 103 | HAPLN1 | 21592 | 11950 | -0.048 | 0.0815 | No | ||
| 104 | EGFL6 | 24006 | 12064 | -0.050 | 0.0762 | No | ||
| 105 | F3 | 15435 | 12137 | -0.052 | 0.0731 | No | ||
| 106 | CXCL10 | 16477 | 12263 | -0.055 | 0.0673 | No | ||
| 107 | SLC15A3 | 23928 | 12373 | -0.058 | 0.0623 | No | ||
| 108 | CYBB | 4579 | 12593 | -0.064 | 0.0514 | No | ||
| 109 | ZBP1 | 14323 | 12749 | -0.069 | 0.0442 | No | ||
| 110 | SERHL | 7570 12713 7571 | 13155 | -0.084 | 0.0236 | No | ||
| 111 | IFIT2 | 23880 | 13429 | -0.095 | 0.0103 | No | ||
| 112 | MAPK10 | 11169 | 13511 | -0.099 | 0.0075 | No | ||
| 113 | STAG2 | 5521 | 13544 | -0.101 | 0.0074 | No | ||
| 114 | GRIA3 | 24349 11996 2649 | 13565 | -0.102 | 0.0079 | No | ||
| 115 | ZNF385 | 22106 2297 | 13595 | -0.104 | 0.0080 | No | ||
| 116 | TCF15 | 14798 | 13767 | -0.113 | 0.0005 | No | ||
| 117 | PRKACA | 18549 3844 | 13814 | -0.116 | -0.0001 | No | ||
| 118 | ETV5 | 22630 | 14139 | -0.137 | -0.0155 | No | ||
| 119 | LGP2 | 8155 | 14159 | -0.138 | -0.0143 | No | ||
| 120 | DUSP10 | 4003 14016 | 14184 | -0.140 | -0.0134 | No | ||
| 121 | FGF9 | 8966 | 14217 | -0.143 | -0.0128 | No | ||
| 122 | ISL1 | 21338 | 14221 | -0.144 | -0.0107 | No | ||
| 123 | ESR1 | 20097 4685 | 14224 | -0.144 | -0.0085 | No | ||
| 124 | IFI35 | 20654 | 14360 | -0.155 | -0.0134 | No | ||
| 125 | ADAM12 | 2207 8545 | 14818 | -0.205 | -0.0348 | No | ||
| 126 | LY86 | 21665 | 14997 | -0.228 | -0.0408 | No | ||
| 127 | SOCS1 | 4522 | 15021 | -0.231 | -0.0384 | No | ||
| 128 | ETV6 | 17264 | 15086 | -0.238 | -0.0381 | No | ||
| 129 | SEMA6D | 2800 14876 90 | 15148 | -0.248 | -0.0375 | No | ||
| 130 | PSMB8 | 5000 | 15261 | -0.265 | -0.0393 | No | ||
| 131 | EOMES | 4669 | 15408 | -0.289 | -0.0426 | No | ||
| 132 | MYB | 3344 3355 19806 | 15442 | -0.297 | -0.0397 | No | ||
| 133 | HOXB4 | 20686 | 15485 | -0.307 | -0.0371 | No | ||
| 134 | CD80 | 22758 | 16016 | -0.435 | -0.0588 | No | ||
| 135 | PSMB10 | 5299 18761 | 16020 | -0.436 | -0.0521 | No | ||
| 136 | ZADH2 | 23501 | 16081 | -0.453 | -0.0481 | No | ||
| 137 | LZTS2 | 10362 | 16172 | -0.479 | -0.0454 | No | ||
| 138 | UBE2L6 | 2768 12169 | 16364 | -0.542 | -0.0471 | No | ||
| 139 | BST2 | 18845 | 16781 | -0.712 | -0.0583 | No | ||
| 140 | ADAM15 | 15275 | 16836 | -0.739 | -0.0494 | No | ||
| 141 | GABARAP | 20815 | 16994 | -0.823 | -0.0448 | No | ||
| 142 | SLC11A2 | 22130 | 17039 | -0.852 | -0.0336 | No | ||
| 143 | CXCR4 | 13844 | 17067 | -0.864 | -0.0214 | No | ||
| 144 | LGALS9 | 1441 20332 1200 | 17101 | -0.878 | -0.0092 | No | ||
| 145 | PSMA3 | 9632 5298 | 17363 | -1.000 | -0.0074 | No | ||
| 146 | PRKAG1 | 22135 | 18092 | -1.496 | -0.0230 | No | ||
| 147 | EXT1 | 22297 | 18114 | -1.522 | 0.0001 | No | ||
| 148 | NT5C3 | 17136 | 18251 | -1.698 | 0.0198 | No |