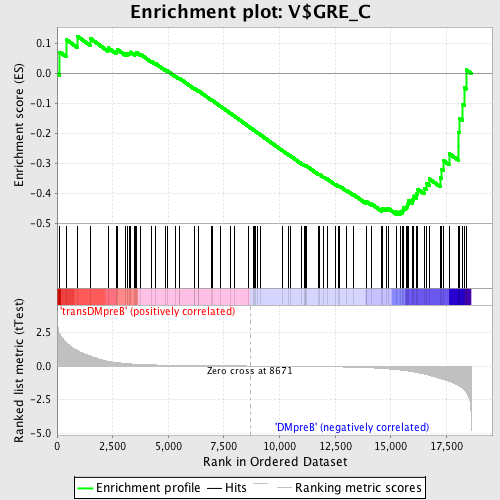
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | V$GRE_C |
| Enrichment Score (ES) | -0.47040588 |
| Normalized Enrichment Score (NES) | -1.2440366 |
| Nominal p-value | 0.097426474 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BTK | 24061 | 118 | 2.378 | 0.0713 | No | ||
| 2 | MEF2C | 3204 9378 | 403 | 1.759 | 0.1135 | No | ||
| 3 | SORL1 | 5474 | 904 | 1.159 | 0.1244 | No | ||
| 4 | ST13 | 12865 | 1500 | 0.738 | 0.1164 | No | ||
| 5 | GPD1 | 22361 | 2290 | 0.357 | 0.0855 | No | ||
| 6 | NOTCH4 | 23277 9475 | 2649 | 0.260 | 0.0747 | No | ||
| 7 | NSD1 | 2134 5197 | 2698 | 0.250 | 0.0803 | No | ||
| 8 | FGF17 | 21757 | 3061 | 0.193 | 0.0670 | No | ||
| 9 | FERD3L | 21292 | 3176 | 0.175 | 0.0666 | No | ||
| 10 | THRAP1 | 20309 | 3250 | 0.165 | 0.0680 | No | ||
| 11 | CDKN2C | 15806 2321 | 3285 | 0.161 | 0.0715 | No | ||
| 12 | MBNL2 | 21932 | 3499 | 0.138 | 0.0645 | No | ||
| 13 | SCNN1A | 17278 | 3537 | 0.135 | 0.0669 | No | ||
| 14 | SFRS6 | 14751 | 3569 | 0.133 | 0.0696 | No | ||
| 15 | LUC7L | 47 1520 | 3762 | 0.117 | 0.0631 | No | ||
| 16 | PAX3 | 13910 | 4251 | 0.087 | 0.0396 | No | ||
| 17 | SEMA3A | 16919 | 4428 | 0.079 | 0.0326 | No | ||
| 18 | ELAVL4 | 15805 4889 9137 | 4859 | 0.063 | 0.0115 | No | ||
| 19 | NPR2 | 16230 | 4955 | 0.060 | 0.0083 | No | ||
| 20 | CDKL5 | 24019 | 5330 | 0.050 | -0.0102 | No | ||
| 21 | SLC7A2 | 4431 8644 | 5478 | 0.046 | -0.0166 | No | ||
| 22 | ANGPTL2 | 15032 18104 | 5519 | 0.046 | -0.0173 | No | ||
| 23 | KIF13A | 21475 | 6156 | 0.033 | -0.0505 | No | ||
| 24 | PRRX1 | 9595 5271 | 6195 | 0.033 | -0.0515 | No | ||
| 25 | ATF7 | 10303 5869 | 6344 | 0.030 | -0.0585 | No | ||
| 26 | TIAL1 | 996 5753 | 6360 | 0.030 | -0.0583 | No | ||
| 27 | CUZD1 | 17598 | 6950 | 0.021 | -0.0894 | No | ||
| 28 | NDUFS1 | 13940 | 6971 | 0.020 | -0.0898 | No | ||
| 29 | PPIL5 | 21255 | 7335 | 0.016 | -0.1089 | No | ||
| 30 | PIK3R1 | 3170 | 7780 | 0.010 | -0.1325 | No | ||
| 31 | LSAMP | 1690 6500 | 7988 | 0.008 | -0.1435 | No | ||
| 32 | KCNA5 | 9203 | 8590 | 0.001 | -0.1759 | No | ||
| 33 | CHST9 | 7727 | 8831 | -0.002 | -0.1887 | No | ||
| 34 | DCX | 24040 8841 4604 | 8883 | -0.002 | -0.1914 | No | ||
| 35 | NRG1 | 3885 | 8910 | -0.003 | -0.1927 | No | ||
| 36 | KCTD15 | 17864 | 9012 | -0.004 | -0.1981 | No | ||
| 37 | RIMS2 | 4370 | 9140 | -0.006 | -0.2047 | No | ||
| 38 | MYL3 | 19291 | 10129 | -0.019 | -0.2574 | No | ||
| 39 | CTGF | 20068 | 10415 | -0.022 | -0.2721 | No | ||
| 40 | BDNF | 14926 2797 | 10474 | -0.023 | -0.2745 | No | ||
| 41 | NNAT | 2764 5180 | 10997 | -0.031 | -0.3016 | No | ||
| 42 | ADK | 3302 3454 8555 | 11121 | -0.033 | -0.3072 | No | ||
| 43 | OTX1 | 20517 | 11153 | -0.033 | -0.3078 | No | ||
| 44 | SMPX | 24220 | 11165 | -0.034 | -0.3073 | No | ||
| 45 | PRDM1 | 19775 3337 | 11223 | -0.034 | -0.3092 | No | ||
| 46 | ROS1 | 19771 | 11761 | -0.044 | -0.3367 | No | ||
| 47 | ONECUT2 | 10346 | 11772 | -0.044 | -0.3358 | No | ||
| 48 | ARX | 24290 | 11981 | -0.048 | -0.3455 | No | ||
| 49 | HOXA3 | 9108 1075 | 12131 | -0.052 | -0.3518 | No | ||
| 50 | LMOD1 | 14119 | 12532 | -0.062 | -0.3714 | No | ||
| 51 | RTKN | 17394 1156 | 12647 | -0.066 | -0.3753 | No | ||
| 52 | LOXL1 | 5002 | 12708 | -0.068 | -0.3764 | No | ||
| 53 | PRG4 | 13592 4036 | 13005 | -0.078 | -0.3898 | No | ||
| 54 | GAB2 | 1821 18184 2025 | 13330 | -0.091 | -0.4043 | No | ||
| 55 | WIF1 | 19866 | 13885 | -0.120 | -0.4303 | No | ||
| 56 | HOXC6 | 22339 2302 | 13907 | -0.121 | -0.4274 | No | ||
| 57 | LHX6 | 9275 2914 | 14128 | -0.136 | -0.4349 | No | ||
| 58 | TCF12 | 10044 3125 | 14586 | -0.178 | -0.4537 | No | ||
| 59 | ATP1A3 | 21 | 14615 | -0.181 | -0.4493 | No | ||
| 60 | SALL2 | 21840 | 14785 | -0.200 | -0.4519 | No | ||
| 61 | MYCT1 | 7572 | 14882 | -0.214 | -0.4501 | No | ||
| 62 | HOXC4 | 4864 | 15252 | -0.263 | -0.4614 | Yes | ||
| 63 | AP4M1 | 3499 16656 | 15420 | -0.292 | -0.4609 | Yes | ||
| 64 | LRRN3 | 21077 | 15543 | -0.317 | -0.4571 | Yes | ||
| 65 | SNX16 | 15374 | 15577 | -0.326 | -0.4482 | Yes | ||
| 66 | ATP5G3 | 14558 | 15683 | -0.351 | -0.4424 | Yes | ||
| 67 | MT2A | 18523 | 15769 | -0.369 | -0.4349 | Yes | ||
| 68 | JPH1 | 13994 | 15799 | -0.377 | -0.4241 | Yes | ||
| 69 | SF3B1 | 13954 | 15970 | -0.423 | -0.4195 | Yes | ||
| 70 | UBE3A | 1209 1084 1830 | 16039 | -0.443 | -0.4087 | Yes | ||
| 71 | LZTS2 | 10362 | 16172 | -0.479 | -0.4002 | Yes | ||
| 72 | EEF1B2 | 4131 12063 | 16213 | -0.489 | -0.3864 | Yes | ||
| 73 | NR2F1 | 7015 21401 | 16502 | -0.589 | -0.3827 | Yes | ||
| 74 | METAP1 | 15154 | 16611 | -0.637 | -0.3677 | Yes | ||
| 75 | CSNK1G1 | 3057 5671 19397 | 16724 | -0.687 | -0.3513 | Yes | ||
| 76 | RAB11A | 7013 | 17231 | -0.939 | -0.3479 | Yes | ||
| 77 | TANK | 2839 15006 2729 | 17296 | -0.975 | -0.3195 | Yes | ||
| 78 | GNB1L | 8921 22827 | 17369 | -1.001 | -0.2907 | Yes | ||
| 79 | RAB10 | 2090 21119 | 17619 | -1.134 | -0.2671 | Yes | ||
| 80 | EGR1 | 23598 | 18043 | -1.444 | -0.2427 | Yes | ||
| 81 | MAPK6 | 19062 | 18044 | -1.445 | -0.1955 | Yes | ||
| 82 | PRKAG1 | 22135 | 18092 | -1.496 | -0.1492 | Yes | ||
| 83 | RRAGD | 11964 | 18227 | -1.664 | -0.1020 | Yes | ||
| 84 | MCM7 | 9372 3568 | 18319 | -1.807 | -0.0479 | Yes | ||
| 85 | TMOD3 | 6945 | 18397 | -1.954 | 0.0118 | Yes |

