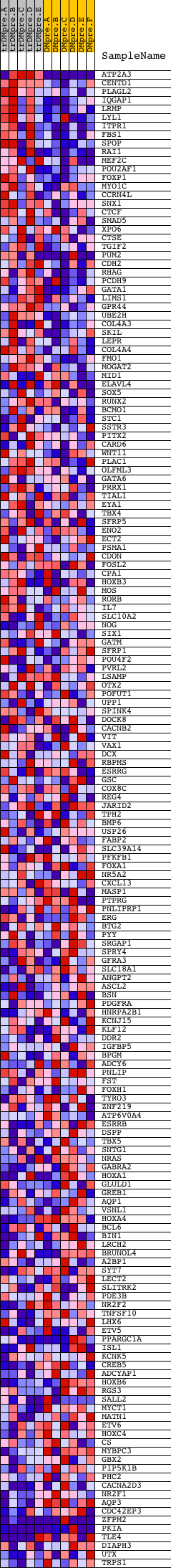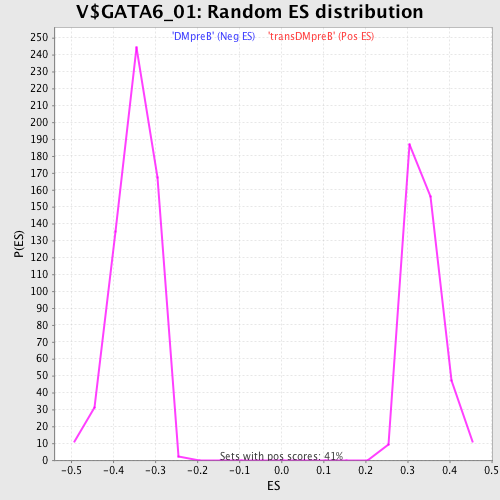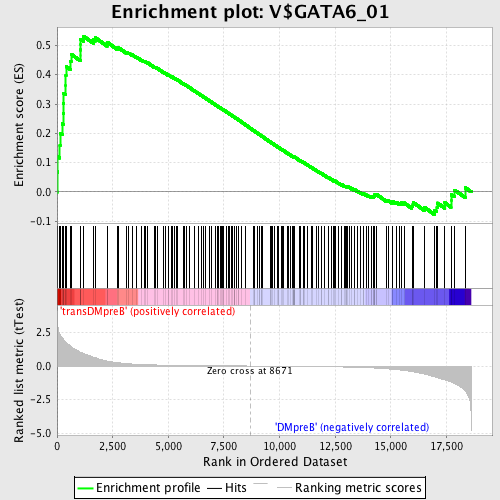
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | V$GATA6_01 |
| Enrichment Score (ES) | 0.53151983 |
| Normalized Enrichment Score (NES) | 1.5754992 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.2517853 |
| FWER p-Value | 0.477 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP2A3 | 20795 1314 | 3 | 3.765 | 0.0686 | Yes | ||
| 2 | CENTD1 | 16529 | 30 | 2.936 | 0.1208 | Yes | ||
| 3 | PLAGL2 | 7055 | 127 | 2.337 | 0.1583 | Yes | ||
| 4 | IQGAP1 | 6619 | 136 | 2.288 | 0.1996 | Yes | ||
| 5 | LRMP | 17244 | 233 | 2.085 | 0.2325 | Yes | ||
| 6 | LYL1 | 18546 | 273 | 2.005 | 0.2670 | Yes | ||
| 7 | ITPR1 | 17341 | 285 | 1.959 | 0.3022 | Yes | ||
| 8 | FBS1 | 18067 | 298 | 1.928 | 0.3367 | Yes | ||
| 9 | SPOP | 5498 | 390 | 1.780 | 0.3643 | Yes | ||
| 10 | RAI1 | 5355 20861 | 394 | 1.778 | 0.3966 | Yes | ||
| 11 | MEF2C | 3204 9378 | 403 | 1.759 | 0.4283 | Yes | ||
| 12 | POU2AF1 | 19455 | 596 | 1.495 | 0.4452 | Yes | ||
| 13 | FOXP1 | 4242 | 632 | 1.442 | 0.4696 | Yes | ||
| 14 | MYO1C | 20777 | 1045 | 1.018 | 0.4658 | Yes | ||
| 15 | CCRN4L | 15599 8708 | 1046 | 1.018 | 0.4844 | Yes | ||
| 16 | SNX1 | 19076 | 1054 | 1.016 | 0.5026 | Yes | ||
| 17 | CTCF | 18490 | 1058 | 1.013 | 0.5209 | Yes | ||
| 18 | SMAD5 | 21621 | 1182 | 0.945 | 0.5315 | Yes | ||
| 19 | XPO6 | 17644 | 1632 | 0.664 | 0.5193 | No | ||
| 20 | CTSE | 8816 | 1717 | 0.618 | 0.5260 | No | ||
| 21 | TGIF2 | 6013 6012 | 2245 | 0.371 | 0.5042 | No | ||
| 22 | PUM2 | 2071 8161 | 2267 | 0.363 | 0.5097 | No | ||
| 23 | CDH2 | 1963 8727 8726 4508 | 2701 | 0.250 | 0.4908 | No | ||
| 24 | RHAG | 1545 1591 23235 | 2767 | 0.238 | 0.4916 | No | ||
| 25 | PCDH9 | 21736 | 3111 | 0.185 | 0.4764 | No | ||
| 26 | GATA1 | 24196 | 3216 | 0.169 | 0.4739 | No | ||
| 27 | LIMS1 | 8493 4290 | 3372 | 0.150 | 0.4682 | No | ||
| 28 | GPR44 | 23926 | 3577 | 0.133 | 0.4596 | No | ||
| 29 | UBE2H | 5823 5822 | 3809 | 0.114 | 0.4491 | No | ||
| 30 | COL4A3 | 8766 | 3905 | 0.108 | 0.4459 | No | ||
| 31 | SKIL | 15617 | 3956 | 0.105 | 0.4451 | No | ||
| 32 | LEPR | 2531 2349 | 4080 | 0.097 | 0.4402 | No | ||
| 33 | COL4A4 | 8767 | 4373 | 0.081 | 0.4259 | No | ||
| 34 | FMO1 | 8976 | 4400 | 0.080 | 0.4259 | No | ||
| 35 | MOGAT2 | 17747 | 4495 | 0.076 | 0.4222 | No | ||
| 36 | MID1 | 5097 5098 | 4780 | 0.065 | 0.4080 | No | ||
| 37 | ELAVL4 | 15805 4889 9137 | 4859 | 0.063 | 0.4049 | No | ||
| 38 | SOX5 | 9850 1044 5480 16937 | 5007 | 0.058 | 0.3980 | No | ||
| 39 | RUNX2 | 4480 8700 | 5008 | 0.058 | 0.3991 | No | ||
| 40 | BCMO1 | 18454 | 5131 | 0.055 | 0.3935 | No | ||
| 41 | STC1 | 21974 | 5167 | 0.054 | 0.3926 | No | ||
| 42 | SSTR3 | 22218 | 5269 | 0.052 | 0.3881 | No | ||
| 43 | PITX2 | 15424 1878 | 5344 | 0.050 | 0.3849 | No | ||
| 44 | CARD6 | 22334 | 5413 | 0.048 | 0.3821 | No | ||
| 45 | WNT11 | 5876 | 5675 | 0.042 | 0.3688 | No | ||
| 46 | PLAC1 | 12079 | 5739 | 0.041 | 0.3661 | No | ||
| 47 | OLFML3 | 15216 | 5803 | 0.040 | 0.3634 | No | ||
| 48 | GATA6 | 96 | 5937 | 0.037 | 0.3569 | No | ||
| 49 | PRRX1 | 9595 5271 | 6195 | 0.033 | 0.3435 | No | ||
| 50 | TIAL1 | 996 5753 | 6360 | 0.030 | 0.3352 | No | ||
| 51 | EYA1 | 4695 4061 | 6495 | 0.028 | 0.3284 | No | ||
| 52 | TBX4 | 5633 | 6570 | 0.027 | 0.3249 | No | ||
| 53 | SFRP5 | 23674 | 6667 | 0.025 | 0.3201 | No | ||
| 54 | ENO2 | 8904 | 6838 | 0.023 | 0.3113 | No | ||
| 55 | ECT2 | 15367 | 6926 | 0.021 | 0.3070 | No | ||
| 56 | PSMA1 | 1627 17669 | 7105 | 0.019 | 0.2977 | No | ||
| 57 | CDON | 12195 | 7208 | 0.017 | 0.2925 | No | ||
| 58 | FOSL2 | 4733 8978 16878 | 7223 | 0.017 | 0.2920 | No | ||
| 59 | CPA1 | 17496 | 7273 | 0.017 | 0.2897 | No | ||
| 60 | HOXB3 | 20685 | 7352 | 0.016 | 0.2857 | No | ||
| 61 | MOS | 16254 | 7388 | 0.015 | 0.2841 | No | ||
| 62 | RORB | 10356 | 7447 | 0.014 | 0.2812 | No | ||
| 63 | IL7 | 4921 | 7471 | 0.014 | 0.2802 | No | ||
| 64 | SLC10A2 | 18939 | 7494 | 0.014 | 0.2793 | No | ||
| 65 | NOG | 13663 | 7615 | 0.012 | 0.2730 | No | ||
| 66 | SIX1 | 9821 | 7711 | 0.011 | 0.2681 | No | ||
| 67 | GATM | 14451 | 7739 | 0.011 | 0.2668 | No | ||
| 68 | SFRP1 | 9805 | 7845 | 0.009 | 0.2613 | No | ||
| 69 | POU4F2 | 9603 5276 | 7870 | 0.009 | 0.2601 | No | ||
| 70 | PVRL2 | 9671 5336 | 7985 | 0.008 | 0.2541 | No | ||
| 71 | LSAMP | 1690 6500 | 7988 | 0.008 | 0.2541 | No | ||
| 72 | OTX2 | 9519 | 7989 | 0.008 | 0.2543 | No | ||
| 73 | POFUT1 | 4697 | 8041 | 0.007 | 0.2516 | No | ||
| 74 | UPP1 | 20947 1385 | 8148 | 0.006 | 0.2460 | No | ||
| 75 | SPINK4 | 16241 | 8150 | 0.006 | 0.2460 | No | ||
| 76 | DOCK8 | 13170 | 8305 | 0.004 | 0.2378 | No | ||
| 77 | CACNB2 | 4466 8678 | 8470 | 0.002 | 0.2289 | No | ||
| 78 | VIT | 23155 | 8809 | -0.002 | 0.2106 | No | ||
| 79 | VAX1 | 23636 | 8882 | -0.002 | 0.2068 | No | ||
| 80 | DCX | 24040 8841 4604 | 8883 | -0.002 | 0.2068 | No | ||
| 81 | RBPMS | 5369 | 9021 | -0.004 | 0.1995 | No | ||
| 82 | ESRRG | 6447 | 9088 | -0.005 | 0.1960 | No | ||
| 83 | GSC | 20990 | 9108 | -0.005 | 0.1950 | No | ||
| 84 | COX8C | 21175 | 9200 | -0.006 | 0.1902 | No | ||
| 85 | REG4 | 15484 | 9207 | -0.006 | 0.1900 | No | ||
| 86 | JARID2 | 9200 | 9218 | -0.007 | 0.1896 | No | ||
| 87 | TPH2 | 19626 | 9570 | -0.011 | 0.1708 | No | ||
| 88 | BMP6 | 8659 | 9612 | -0.012 | 0.1687 | No | ||
| 89 | USP26 | 24155 | 9629 | -0.012 | 0.1681 | No | ||
| 90 | FABP2 | 15430 | 9667 | -0.013 | 0.1663 | No | ||
| 91 | SLC39A14 | 3517 5611 21762 | 9775 | -0.014 | 0.1608 | No | ||
| 92 | PFKFB1 | 9552 | 9909 | -0.016 | 0.1538 | No | ||
| 93 | FOXA1 | 21060 | 9938 | -0.016 | 0.1526 | No | ||
| 94 | NR5A2 | 4105 13817 | 9942 | -0.016 | 0.1528 | No | ||
| 95 | CXCL13 | 16789 | 10074 | -0.018 | 0.1460 | No | ||
| 96 | MASP1 | 1632 22626 | 10115 | -0.018 | 0.1441 | No | ||
| 97 | PTPRG | 22097 3151 | 10187 | -0.019 | 0.1406 | No | ||
| 98 | PNLIPRP1 | 23835 | 10336 | -0.021 | 0.1330 | No | ||
| 99 | ERG | 1686 8915 | 10402 | -0.022 | 0.1299 | No | ||
| 100 | BTG2 | 13832 | 10492 | -0.024 | 0.1255 | No | ||
| 101 | PYY | 20207 | 10558 | -0.025 | 0.1224 | No | ||
| 102 | SRGAP1 | 19615 | 10602 | -0.025 | 0.1206 | No | ||
| 103 | SPRY4 | 23448 | 10636 | -0.026 | 0.1192 | No | ||
| 104 | GFRA3 | 23469 | 10646 | -0.026 | 0.1192 | No | ||
| 105 | SLC18A1 | 18862 | 10651 | -0.026 | 0.1195 | No | ||
| 106 | ANGPT2 | 14885 3783 18923 | 10662 | -0.026 | 0.1194 | No | ||
| 107 | ASCL2 | 5079 | 10681 | -0.026 | 0.1189 | No | ||
| 108 | BSN | 18997 | 10883 | -0.029 | 0.1085 | No | ||
| 109 | PDGFRA | 16824 | 10927 | -0.030 | 0.1068 | No | ||
| 110 | HNRPA2B1 | 1187 7001 7000 1040 | 10954 | -0.030 | 0.1059 | No | ||
| 111 | KCNJ15 | 4943 9208 | 11070 | -0.032 | 0.1002 | No | ||
| 112 | KLF12 | 4960 2637 21732 9228 | 11071 | -0.032 | 0.1008 | No | ||
| 113 | DDR2 | 13767 | 11111 | -0.033 | 0.0993 | No | ||
| 114 | IGFBP5 | 4899 | 11262 | -0.035 | 0.0918 | No | ||
| 115 | BPGM | 17489 | 11437 | -0.038 | 0.0831 | No | ||
| 116 | ADCY6 | 22139 2283 8551 | 11490 | -0.039 | 0.0810 | No | ||
| 117 | PNLIP | 23830 | 11656 | -0.042 | 0.0728 | No | ||
| 118 | FST | 8985 | 11763 | -0.044 | 0.0678 | No | ||
| 119 | FOXH1 | 22238 | 11877 | -0.046 | 0.0626 | No | ||
| 120 | TYRO3 | 5811 | 11890 | -0.047 | 0.0628 | No | ||
| 121 | ZNF219 | 12835 | 12021 | -0.049 | 0.0566 | No | ||
| 122 | ATP6V0A4 | 8934 | 12189 | -0.054 | 0.0485 | No | ||
| 123 | ESRRB | 21196 | 12202 | -0.054 | 0.0489 | No | ||
| 124 | DSPP | 8867 | 12349 | -0.057 | 0.0420 | No | ||
| 125 | TBX5 | 3531 5634 | 12442 | -0.060 | 0.0381 | No | ||
| 126 | SNTG1 | 7717 | 12445 | -0.060 | 0.0391 | No | ||
| 127 | NRAS | 5191 | 12495 | -0.061 | 0.0376 | No | ||
| 128 | GABRA2 | 4746 | 12664 | -0.067 | 0.0297 | No | ||
| 129 | HOXA1 | 1005 17152 | 12786 | -0.071 | 0.0244 | No | ||
| 130 | GLULD1 | 14285 | 12794 | -0.071 | 0.0253 | No | ||
| 131 | GREB1 | 6490 2152 | 12931 | -0.075 | 0.0193 | No | ||
| 132 | AQP1 | 1057 17437 | 12940 | -0.076 | 0.0203 | No | ||
| 133 | VSNL1 | 6527 | 13009 | -0.078 | 0.0180 | No | ||
| 134 | HOXA4 | 17148 | 13012 | -0.079 | 0.0193 | No | ||
| 135 | BCL6 | 22624 | 13041 | -0.079 | 0.0193 | No | ||
| 136 | BIN1 | 6634 2017 | 13135 | -0.083 | 0.0157 | No | ||
| 137 | LRCH2 | 24035 | 13248 | -0.087 | 0.0113 | No | ||
| 138 | BRUNOL4 | 2010 4229 | 13352 | -0.091 | 0.0073 | No | ||
| 139 | A2BP1 | 11212 1652 1689 | 13356 | -0.092 | 0.0089 | No | ||
| 140 | SYT7 | 3670 3698 23932 | 13513 | -0.100 | 0.0022 | No | ||
| 141 | LECT2 | 21443 | 13653 | -0.107 | -0.0034 | No | ||
| 142 | SLITRK2 | 24322 | 13765 | -0.113 | -0.0073 | No | ||
| 143 | PDE3B | 5236 | 13786 | -0.114 | -0.0063 | No | ||
| 144 | NR2F2 | 8619 409 | 13896 | -0.121 | -0.0100 | No | ||
| 145 | TNFSF10 | 15626 | 14007 | -0.128 | -0.0137 | No | ||
| 146 | LHX6 | 9275 2914 | 14128 | -0.136 | -0.0177 | No | ||
| 147 | ETV5 | 22630 | 14139 | -0.137 | -0.0157 | No | ||
| 148 | PPARGC1A | 16533 | 14203 | -0.142 | -0.0166 | No | ||
| 149 | ISL1 | 21338 | 14221 | -0.144 | -0.0149 | No | ||
| 150 | KCNK5 | 4946 9212 | 14249 | -0.146 | -0.0137 | No | ||
| 151 | CREB5 | 10551 | 14255 | -0.146 | -0.0113 | No | ||
| 152 | ADCYAP1 | 4348 | 14281 | -0.148 | -0.0099 | No | ||
| 153 | HOXB6 | 20688 | 14339 | -0.153 | -0.0102 | No | ||
| 154 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 14377 | -0.157 | -0.0094 | No | ||
| 155 | SALL2 | 21840 | 14785 | -0.200 | -0.0278 | No | ||
| 156 | MYCT1 | 7572 | 14882 | -0.214 | -0.0291 | No | ||
| 157 | MATN1 | 16060 | 15063 | -0.235 | -0.0345 | No | ||
| 158 | ETV6 | 17264 | 15086 | -0.238 | -0.0314 | No | ||
| 159 | HOXC4 | 4864 | 15252 | -0.263 | -0.0355 | No | ||
| 160 | CS | 19839 | 15391 | -0.287 | -0.0378 | No | ||
| 161 | MYBPC3 | 9434 | 15479 | -0.305 | -0.0369 | No | ||
| 162 | GBX2 | 13887 | 15598 | -0.331 | -0.0373 | No | ||
| 163 | PIP5K1B | 5251 1841 | 15951 | -0.416 | -0.0488 | No | ||
| 164 | PHC2 | 16075 | 15994 | -0.430 | -0.0432 | No | ||
| 165 | CACNA2D3 | 8677 | 15997 | -0.430 | -0.0354 | No | ||
| 166 | NR2F1 | 7015 21401 | 16502 | -0.589 | -0.0520 | No | ||
| 167 | AQP3 | 15915 | 16960 | -0.804 | -0.0621 | No | ||
| 168 | CDC42EP3 | 6436 | 17074 | -0.867 | -0.0524 | No | ||
| 169 | ZFPM2 | 22483 | 17097 | -0.876 | -0.0376 | No | ||
| 170 | PKIA | 15639 | 17428 | -1.023 | -0.0368 | No | ||
| 171 | TLE4 | 23720 3697 | 17706 | -1.183 | -0.0302 | No | ||
| 172 | DIAPH3 | 7117 | 17711 | -1.189 | -0.0087 | No | ||
| 173 | UTX | 10266 2574 | 17866 | -1.300 | 0.0067 | No | ||
| 174 | TRPS1 | 8195 | 18344 | -1.859 | 0.0147 | No |