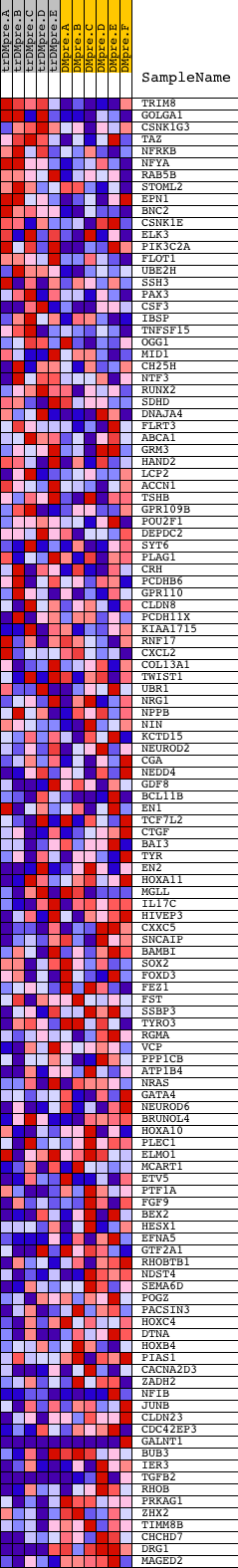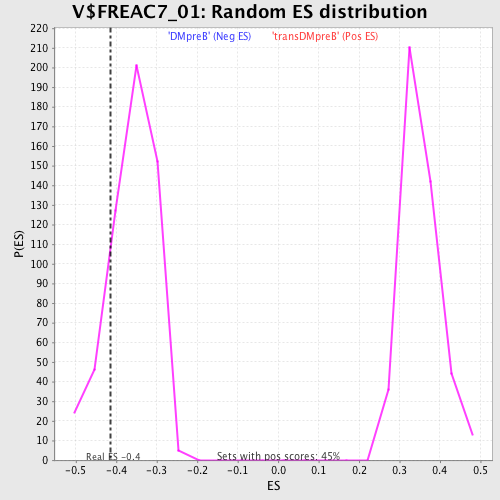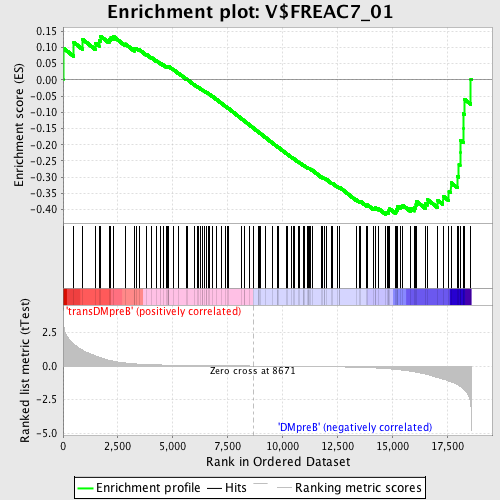
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | V$FREAC7_01 |
| Enrichment Score (ES) | -0.41488892 |
| Normalized Enrichment Score (NES) | -1.1483755 |
| Nominal p-value | 0.16396396 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TRIM8 | 23824 | 5 | 3.715 | 0.0980 | No | ||
| 2 | GOLGA1 | 14595 | 488 | 1.641 | 0.1154 | No | ||
| 3 | CSNK1G3 | 12873 | 892 | 1.169 | 0.1245 | No | ||
| 4 | TAZ | 12415 | 1487 | 0.750 | 0.1122 | No | ||
| 5 | NFRKB | 10642 | 1642 | 0.660 | 0.1213 | No | ||
| 6 | NFYA | 5172 | 1703 | 0.626 | 0.1347 | No | ||
| 7 | RAB5B | 359 19591 3430 | 2096 | 0.421 | 0.1246 | No | ||
| 8 | STOML2 | 15902 | 2180 | 0.388 | 0.1304 | No | ||
| 9 | EPN1 | 18401 | 2291 | 0.357 | 0.1339 | No | ||
| 10 | BNC2 | 15850 | 2836 | 0.228 | 0.1105 | No | ||
| 11 | CSNK1E | 6570 2211 11332 | 3242 | 0.166 | 0.0930 | No | ||
| 12 | ELK3 | 4663 94 3388 | 3251 | 0.165 | 0.0969 | No | ||
| 13 | PIK3C2A | 17667 | 3326 | 0.156 | 0.0970 | No | ||
| 14 | FLOT1 | 23249 1613 | 3461 | 0.141 | 0.0935 | No | ||
| 15 | UBE2H | 5823 5822 | 3809 | 0.114 | 0.0778 | No | ||
| 16 | SSH3 | 3709 10890 | 4039 | 0.100 | 0.0680 | No | ||
| 17 | PAX3 | 13910 | 4251 | 0.087 | 0.0589 | No | ||
| 18 | CSF3 | 1394 20671 | 4443 | 0.078 | 0.0506 | No | ||
| 19 | IBSP | 16775 | 4564 | 0.073 | 0.0461 | No | ||
| 20 | TNFSF15 | 15863 | 4710 | 0.068 | 0.0400 | No | ||
| 21 | OGG1 | 17331 | 4763 | 0.066 | 0.0390 | No | ||
| 22 | MID1 | 5097 5098 | 4780 | 0.065 | 0.0398 | No | ||
| 23 | CH25H | 23695 | 4813 | 0.065 | 0.0398 | No | ||
| 24 | NTF3 | 16991 | 4816 | 0.064 | 0.0414 | No | ||
| 25 | RUNX2 | 4480 8700 | 5008 | 0.058 | 0.0326 | No | ||
| 26 | SDHD | 19125 | 5278 | 0.051 | 0.0194 | No | ||
| 27 | DNAJA4 | 19446 | 5607 | 0.044 | 0.0029 | No | ||
| 28 | FLRT3 | 7730 12930 2673 | 5651 | 0.043 | 0.0017 | No | ||
| 29 | ABCA1 | 15881 | 5965 | 0.036 | -0.0143 | No | ||
| 30 | GRM3 | 16929 | 6116 | 0.034 | -0.0215 | No | ||
| 31 | HAND2 | 18615 | 6152 | 0.033 | -0.0225 | No | ||
| 32 | LCP2 | 4988 9268 | 6264 | 0.031 | -0.0277 | No | ||
| 33 | ACCN1 | 20327 1194 | 6365 | 0.030 | -0.0323 | No | ||
| 34 | TSHB | 15218 | 6369 | 0.030 | -0.0317 | No | ||
| 35 | GPR109B | 16374 | 6435 | 0.029 | -0.0344 | No | ||
| 36 | POU2F1 | 5275 3989 4065 4010 | 6540 | 0.027 | -0.0394 | No | ||
| 37 | DEPDC2 | 14295 | 6543 | 0.027 | -0.0387 | No | ||
| 38 | SYT6 | 13437 7042 12040 | 6616 | 0.026 | -0.0420 | No | ||
| 39 | PLAG1 | 7148 15949 12161 | 6686 | 0.025 | -0.0450 | No | ||
| 40 | CRH | 8784 | 6825 | 0.023 | -0.0519 | No | ||
| 41 | PCDHB6 | 8224 | 6998 | 0.020 | -0.0607 | No | ||
| 42 | GPR110 | 23234 | 7221 | 0.017 | -0.0722 | No | ||
| 43 | CLDN8 | 22549 1747 | 7386 | 0.015 | -0.0807 | No | ||
| 44 | PCDH11X | 2559 | 7490 | 0.014 | -0.0859 | No | ||
| 45 | KIAA1715 | 7629 | 7526 | 0.013 | -0.0874 | No | ||
| 46 | RNF17 | 11389 | 8124 | 0.006 | -0.1196 | No | ||
| 47 | CXCL2 | 9790 | 8251 | 0.005 | -0.1262 | No | ||
| 48 | COL13A1 | 8763 | 8288 | 0.005 | -0.1281 | No | ||
| 49 | TWIST1 | 21291 | 8501 | 0.002 | -0.1395 | No | ||
| 50 | UBR1 | 5828 | 8690 | -0.000 | -0.1496 | No | ||
| 51 | NRG1 | 3885 | 8910 | -0.003 | -0.1614 | No | ||
| 52 | NPPB | 15994 | 8924 | -0.003 | -0.1620 | No | ||
| 53 | NIN | 9463 2103 | 8964 | -0.003 | -0.1641 | No | ||
| 54 | KCTD15 | 17864 | 9012 | -0.004 | -0.1665 | No | ||
| 55 | NEUROD2 | 20262 | 9204 | -0.006 | -0.1766 | No | ||
| 56 | CGA | 16246 | 9520 | -0.011 | -0.1934 | No | ||
| 57 | NEDD4 | 19384 3101 | 9763 | -0.014 | -0.2061 | No | ||
| 58 | GDF8 | 9411 | 9791 | -0.014 | -0.2072 | No | ||
| 59 | BCL11B | 7190 | 9826 | -0.015 | -0.2087 | No | ||
| 60 | EN1 | 387 14155 | 10178 | -0.019 | -0.2271 | No | ||
| 61 | TCF7L2 | 10048 5646 | 10216 | -0.020 | -0.2286 | No | ||
| 62 | CTGF | 20068 | 10415 | -0.022 | -0.2387 | No | ||
| 63 | BAI3 | 5580 | 10504 | -0.024 | -0.2429 | No | ||
| 64 | TYR | 17763 | 10535 | -0.024 | -0.2438 | No | ||
| 65 | EN2 | 16898 | 10730 | -0.027 | -0.2536 | No | ||
| 66 | HOXA11 | 17146 | 10778 | -0.028 | -0.2554 | No | ||
| 67 | MGLL | 17368 1145 | 10934 | -0.030 | -0.2630 | No | ||
| 68 | IL17C | 18438 | 10951 | -0.030 | -0.2631 | No | ||
| 69 | HIVEP3 | 4973 | 11020 | -0.031 | -0.2659 | No | ||
| 70 | CXXC5 | 12523 | 11134 | -0.033 | -0.2712 | No | ||
| 71 | SNCAIP | 12589 | 11176 | -0.034 | -0.2725 | No | ||
| 72 | BAMBI | 23629 | 11188 | -0.034 | -0.2722 | No | ||
| 73 | SOX2 | 9849 15612 | 11229 | -0.034 | -0.2735 | No | ||
| 74 | FOXD3 | 9088 | 11289 | -0.035 | -0.2757 | No | ||
| 75 | FEZ1 | 19520 3100 | 11346 | -0.036 | -0.2778 | No | ||
| 76 | FST | 8985 | 11763 | -0.044 | -0.2991 | No | ||
| 77 | SSBP3 | 2439 7814 2431 | 11832 | -0.046 | -0.3016 | No | ||
| 78 | TYRO3 | 5811 | 11890 | -0.047 | -0.3034 | No | ||
| 79 | RGMA | 18213 | 11898 | -0.047 | -0.3026 | No | ||
| 80 | VCP | 11254 | 12012 | -0.049 | -0.3074 | No | ||
| 81 | PPP1CB | 5282 3529 3504 | 12239 | -0.055 | -0.3182 | No | ||
| 82 | ATP1B4 | 12586 | 12284 | -0.056 | -0.3191 | No | ||
| 83 | NRAS | 5191 | 12495 | -0.061 | -0.3288 | No | ||
| 84 | GATA4 | 21792 4755 | 12595 | -0.064 | -0.3325 | No | ||
| 85 | NEUROD6 | 17138 | 12610 | -0.065 | -0.3315 | No | ||
| 86 | BRUNOL4 | 2010 4229 | 13352 | -0.091 | -0.3691 | No | ||
| 87 | HOXA10 | 17145 989 | 13519 | -0.100 | -0.3755 | No | ||
| 88 | PLEC1 | 2273 2198 2205 2178 2230 22247 2213 2231 2295 2172 5263 2266 | 13573 | -0.103 | -0.3756 | No | ||
| 89 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 13839 | -0.118 | -0.3868 | No | ||
| 90 | MCART1 | 10492 | 13868 | -0.119 | -0.3852 | No | ||
| 91 | ETV5 | 22630 | 14139 | -0.137 | -0.3962 | No | ||
| 92 | PTF1A | 15110 | 14215 | -0.143 | -0.3964 | No | ||
| 93 | FGF9 | 8966 | 14217 | -0.143 | -0.3927 | No | ||
| 94 | BEX2 | 24055 | 14376 | -0.157 | -0.3971 | No | ||
| 95 | HESX1 | 22070 | 14706 | -0.191 | -0.4098 | Yes | ||
| 96 | EFNA5 | 4655 8884 | 14762 | -0.198 | -0.4076 | Yes | ||
| 97 | GTF2A1 | 2064 8187 8188 13512 | 14827 | -0.206 | -0.4056 | Yes | ||
| 98 | RHOBTB1 | 12789 | 14848 | -0.209 | -0.4011 | Yes | ||
| 99 | NDST4 | 12259 7228 | 14890 | -0.215 | -0.3977 | Yes | ||
| 100 | SEMA6D | 2800 14876 90 | 15148 | -0.248 | -0.4050 | Yes | ||
| 101 | POGZ | 6031 1911 | 15192 | -0.255 | -0.4006 | Yes | ||
| 102 | PACSIN3 | 14948 8149 2776 2665 | 15236 | -0.261 | -0.3960 | Yes | ||
| 103 | HOXC4 | 4864 | 15252 | -0.263 | -0.3899 | Yes | ||
| 104 | DTNA | 8870 4647 23611 1991 | 15379 | -0.285 | -0.3892 | Yes | ||
| 105 | HOXB4 | 20686 | 15485 | -0.307 | -0.3867 | Yes | ||
| 106 | PIAS1 | 7126 | 15828 | -0.383 | -0.3951 | Yes | ||
| 107 | CACNA2D3 | 8677 | 15997 | -0.430 | -0.3928 | Yes | ||
| 108 | ZADH2 | 23501 | 16081 | -0.453 | -0.3853 | Yes | ||
| 109 | NFIB | 15855 | 16096 | -0.456 | -0.3740 | Yes | ||
| 110 | JUNB | 9201 | 16520 | -0.594 | -0.3811 | Yes | ||
| 111 | CLDN23 | 18893 | 16608 | -0.636 | -0.3690 | Yes | ||
| 112 | CDC42EP3 | 6436 | 17074 | -0.867 | -0.3712 | Yes | ||
| 113 | GALNT1 | 8998 23609 8999 4750 | 17314 | -0.984 | -0.3581 | Yes | ||
| 114 | BUB3 | 18045 | 17586 | -1.118 | -0.3432 | Yes | ||
| 115 | IER3 | 23248 | 17679 | -1.169 | -0.3173 | Yes | ||
| 116 | TGFB2 | 5743 | 17974 | -1.384 | -0.2965 | Yes | ||
| 117 | RHOB | 4411 | 18042 | -1.444 | -0.2620 | Yes | ||
| 118 | PRKAG1 | 22135 | 18092 | -1.496 | -0.2250 | Yes | ||
| 119 | ZHX2 | 11842 | 18096 | -1.501 | -0.1855 | Yes | ||
| 120 | TIMM8B | 11391 | 18239 | -1.678 | -0.1488 | Yes | ||
| 121 | CHCHD7 | 16280 | 18243 | -1.684 | -0.1044 | Yes | ||
| 122 | DRG1 | 1199 20553 | 18305 | -1.786 | -0.0605 | Yes | ||
| 123 | MAGED2 | 24034 | 18586 | -2.920 | 0.0016 | Yes |