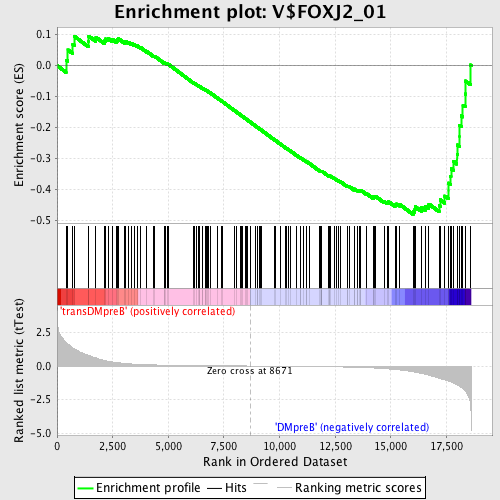
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | V$FOXJ2_01 |
| Enrichment Score (ES) | -0.48205453 |
| Normalized Enrichment Score (NES) | -1.3451972 |
| Nominal p-value | 0.028268551 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MEF2C | 3204 9378 | 403 | 1.759 | 0.0180 | No | ||
| 2 | GOLGA1 | 14595 | 488 | 1.641 | 0.0506 | No | ||
| 3 | KLF3 | 4961 | 707 | 1.357 | 0.0694 | No | ||
| 4 | PCF11 | 121 | 771 | 1.282 | 0.0950 | No | ||
| 5 | ATP2A2 | 4421 3481 | 1416 | 0.790 | 0.0781 | No | ||
| 6 | BRIP1 | 20311 | 1424 | 0.782 | 0.0954 | No | ||
| 7 | PHTF2 | 16599 | 1744 | 0.601 | 0.0917 | No | ||
| 8 | CPNE1 | 11186 | 2135 | 0.406 | 0.0798 | No | ||
| 9 | STOML2 | 15902 | 2180 | 0.388 | 0.0862 | No | ||
| 10 | ACACA | 309 4209 | 2298 | 0.356 | 0.0879 | No | ||
| 11 | HNF4A | 14746 | 2467 | 0.301 | 0.0856 | No | ||
| 12 | CUTL1 | 3507 3480 8820 3609 4577 | 2665 | 0.257 | 0.0808 | No | ||
| 13 | RPS6KB1 | 7815 1207 13040 | 2699 | 0.250 | 0.0846 | No | ||
| 14 | CD68 | 4501 | 2752 | 0.241 | 0.0873 | No | ||
| 15 | FOXP2 | 4312 17523 8523 | 3032 | 0.196 | 0.0766 | No | ||
| 16 | FEN1 | 8961 | 3090 | 0.188 | 0.0778 | No | ||
| 17 | PDE7A | 9544 1788 | 3212 | 0.170 | 0.0751 | No | ||
| 18 | PIK3C2A | 17667 | 3326 | 0.156 | 0.0725 | No | ||
| 19 | FLOT1 | 23249 1613 | 3461 | 0.141 | 0.0684 | No | ||
| 20 | PVRL1 | 3006 19482 12211 7192 | 3595 | 0.131 | 0.0642 | No | ||
| 21 | LUC7L | 47 1520 | 3762 | 0.117 | 0.0579 | No | ||
| 22 | SSH3 | 3709 10890 | 4039 | 0.100 | 0.0452 | No | ||
| 23 | NCAM1 | 5149 | 4347 | 0.082 | 0.0304 | No | ||
| 24 | HOXA7 | 4862 | 4378 | 0.081 | 0.0306 | No | ||
| 25 | CH25H | 23695 | 4813 | 0.065 | 0.0086 | No | ||
| 26 | NTF3 | 16991 | 4816 | 0.064 | 0.0100 | No | ||
| 27 | LGI1 | 23867 | 4891 | 0.062 | 0.0074 | No | ||
| 28 | LTA | 23003 | 4950 | 0.060 | 0.0056 | No | ||
| 29 | RUNX2 | 4480 8700 | 5008 | 0.058 | 0.0038 | No | ||
| 30 | GRM3 | 16929 | 6116 | 0.034 | -0.0553 | No | ||
| 31 | HAND2 | 18615 | 6152 | 0.033 | -0.0564 | No | ||
| 32 | LCP2 | 4988 9268 | 6264 | 0.031 | -0.0617 | No | ||
| 33 | CYP26B1 | 17093 | 6349 | 0.030 | -0.0656 | No | ||
| 34 | EGR2 | 8886 | 6380 | 0.030 | -0.0665 | No | ||
| 35 | TNF | 23004 | 6399 | 0.029 | -0.0668 | No | ||
| 36 | DEPDC2 | 14295 | 6543 | 0.027 | -0.0740 | No | ||
| 37 | FOXB1 | 12254 19069 | 6547 | 0.027 | -0.0735 | No | ||
| 38 | HEY2 | 19794 | 6650 | 0.025 | -0.0785 | No | ||
| 39 | PLAG1 | 7148 15949 12161 | 6686 | 0.025 | -0.0798 | No | ||
| 40 | ANGPT1 | 8588 2260 22303 | 6697 | 0.025 | -0.0798 | No | ||
| 41 | NTN1 | 20400 | 6758 | 0.024 | -0.0825 | No | ||
| 42 | CRH | 8784 | 6825 | 0.023 | -0.0856 | No | ||
| 43 | SLN | 19450 | 6877 | 0.022 | -0.0878 | No | ||
| 44 | IMPDH1 | 17197 1131 | 7187 | 0.018 | -0.1041 | No | ||
| 45 | CLDN8 | 22549 1747 | 7386 | 0.015 | -0.1145 | No | ||
| 46 | REST | 16818 | 7430 | 0.015 | -0.1165 | No | ||
| 47 | OTX2 | 9519 | 7989 | 0.008 | -0.1465 | No | ||
| 48 | ARTN | 15783 2430 2532 | 8070 | 0.007 | -0.1507 | No | ||
| 49 | CXCL2 | 9790 | 8251 | 0.005 | -0.1603 | No | ||
| 50 | COL13A1 | 8763 | 8288 | 0.005 | -0.1622 | No | ||
| 51 | TNR | 5787 | 8354 | 0.004 | -0.1656 | No | ||
| 52 | RGS6 | 21219 | 8457 | 0.003 | -0.1711 | No | ||
| 53 | TWIST1 | 21291 | 8501 | 0.002 | -0.1733 | No | ||
| 54 | TBXAS1 | 17480 1105 | 8565 | 0.001 | -0.1767 | No | ||
| 55 | UBR1 | 5828 | 8690 | -0.000 | -0.1834 | No | ||
| 56 | E2F5 | 15633 | 8699 | -0.000 | -0.1838 | No | ||
| 57 | NRG1 | 3885 | 8910 | -0.003 | -0.1951 | No | ||
| 58 | BUB1 | 8665 | 9007 | -0.004 | -0.2003 | No | ||
| 59 | KCTD15 | 17864 | 9012 | -0.004 | -0.2004 | No | ||
| 60 | ESRRG | 6447 | 9088 | -0.005 | -0.2043 | No | ||
| 61 | MITF | 17349 | 9128 | -0.005 | -0.2063 | No | ||
| 62 | NEUROD2 | 20262 | 9204 | -0.006 | -0.2102 | No | ||
| 63 | GDF8 | 9411 | 9791 | -0.014 | -0.2416 | No | ||
| 64 | BCL11B | 7190 | 9826 | -0.015 | -0.2431 | No | ||
| 65 | FOXN1 | 20337 | 10054 | -0.018 | -0.2550 | No | ||
| 66 | CHN2 | 7652 | 10287 | -0.021 | -0.2671 | No | ||
| 67 | RORA | 3019 5388 | 10305 | -0.021 | -0.2675 | No | ||
| 68 | SOX6 | 3684 5481 9851 | 10394 | -0.022 | -0.2718 | No | ||
| 69 | BAI3 | 5580 | 10504 | -0.024 | -0.2772 | No | ||
| 70 | HOXA11 | 17146 | 10778 | -0.028 | -0.2913 | No | ||
| 71 | MGLL | 17368 1145 | 10934 | -0.030 | -0.2990 | No | ||
| 72 | IL17C | 18438 | 10951 | -0.030 | -0.2992 | No | ||
| 73 | KLF12 | 4960 2637 21732 9228 | 11071 | -0.032 | -0.3049 | No | ||
| 74 | BAMBI | 23629 | 11188 | -0.034 | -0.3104 | No | ||
| 75 | PRDM1 | 19775 3337 | 11223 | -0.034 | -0.3115 | No | ||
| 76 | TWIST2 | 14185 | 11339 | -0.036 | -0.3169 | No | ||
| 77 | GNAO1 | 4785 3829 | 11790 | -0.045 | -0.3402 | No | ||
| 78 | DRD3 | 22750 | 11837 | -0.046 | -0.3417 | No | ||
| 79 | TYRO3 | 5811 | 11890 | -0.047 | -0.3435 | No | ||
| 80 | SLIT3 | 20925 | 11896 | -0.047 | -0.3427 | No | ||
| 81 | RGMA | 18213 | 11898 | -0.047 | -0.3417 | No | ||
| 82 | DMD | 24295 2647 | 12193 | -0.054 | -0.3563 | No | ||
| 83 | ANGPTL1 | 10195 8589 13064 | 12238 | -0.055 | -0.3575 | No | ||
| 84 | PPP1CB | 5282 3529 3504 | 12239 | -0.055 | -0.3563 | No | ||
| 85 | ATP1B4 | 12586 | 12284 | -0.056 | -0.3574 | No | ||
| 86 | SLC35C2 | 10463 14339 | 12464 | -0.061 | -0.3657 | No | ||
| 87 | SLC39A8 | 12547 1776 | 12563 | -0.063 | -0.3696 | No | ||
| 88 | IRS4 | 9183 4926 | 12661 | -0.067 | -0.3733 | No | ||
| 89 | GAP43 | 9001 | 12747 | -0.069 | -0.3763 | No | ||
| 90 | FABP4 | 8601 | 13035 | -0.079 | -0.3901 | No | ||
| 91 | TRERF1 | 1546 23213 | 13068 | -0.080 | -0.3900 | No | ||
| 92 | ORC4L | 11172 6460 | 13121 | -0.082 | -0.3909 | No | ||
| 93 | BRUNOL4 | 2010 4229 | 13352 | -0.091 | -0.4013 | No | ||
| 94 | A2BP1 | 11212 1652 1689 | 13356 | -0.092 | -0.3994 | No | ||
| 95 | GFI1 | 16451 | 13485 | -0.098 | -0.4041 | No | ||
| 96 | HOXA10 | 17145 989 | 13519 | -0.100 | -0.4036 | No | ||
| 97 | PLEC1 | 2273 2198 2205 2178 2230 22247 2213 2231 2295 2172 5263 2266 | 13573 | -0.103 | -0.4042 | No | ||
| 98 | IFNA5 | 9151 | 13628 | -0.106 | -0.4047 | No | ||
| 99 | DAAM1 | 5536 | 13639 | -0.106 | -0.4028 | No | ||
| 100 | HOXC6 | 22339 2302 | 13907 | -0.121 | -0.4145 | No | ||
| 101 | PTF1A | 15110 | 14215 | -0.143 | -0.4279 | No | ||
| 102 | FGF9 | 8966 | 14217 | -0.143 | -0.4247 | No | ||
| 103 | CREB5 | 10551 | 14255 | -0.146 | -0.4234 | No | ||
| 104 | POLR3F | 2814 7674 | 14314 | -0.151 | -0.4231 | No | ||
| 105 | HESX1 | 22070 | 14706 | -0.191 | -0.4400 | No | ||
| 106 | SCG3 | 19061 | 14841 | -0.208 | -0.4425 | No | ||
| 107 | NCDN | 15756 2487 6471 | 14904 | -0.217 | -0.4410 | No | ||
| 108 | POGZ | 6031 1911 | 15192 | -0.255 | -0.4507 | No | ||
| 109 | HOXC4 | 4864 | 15252 | -0.263 | -0.4480 | No | ||
| 110 | DTNA | 8870 4647 23611 1991 | 15379 | -0.285 | -0.4483 | No | ||
| 111 | GNAZ | 71 | 16003 | -0.431 | -0.4723 | Yes | ||
| 112 | ZADH2 | 23501 | 16081 | -0.453 | -0.4662 | Yes | ||
| 113 | NFIB | 15855 | 16096 | -0.456 | -0.4567 | Yes | ||
| 114 | DNAJB12 | 7146 | 16386 | -0.548 | -0.4599 | Yes | ||
| 115 | RAB3IP | 19623 | 16545 | -0.609 | -0.4547 | Yes | ||
| 116 | FSTL1 | 22765 | 16709 | -0.678 | -0.4482 | Yes | ||
| 117 | PPM1D | 20721 | 17172 | -0.905 | -0.4527 | Yes | ||
| 118 | HSF2 | 9128 | 17223 | -0.934 | -0.4343 | Yes | ||
| 119 | PKIA | 15639 | 17428 | -1.023 | -0.4222 | Yes | ||
| 120 | BUB3 | 18045 | 17586 | -1.118 | -0.4054 | Yes | ||
| 121 | NEO1 | 5161 | 17587 | -1.118 | -0.3801 | Yes | ||
| 122 | IER3 | 23248 | 17679 | -1.169 | -0.3586 | Yes | ||
| 123 | TLE4 | 23720 3697 | 17706 | -1.183 | -0.3332 | Yes | ||
| 124 | ACP6 | 12394 | 17828 | -1.272 | -0.3110 | Yes | ||
| 125 | TGFB2 | 5743 | 17974 | -1.384 | -0.2875 | Yes | ||
| 126 | ICK | 7135 12146 | 17995 | -1.401 | -0.2569 | Yes | ||
| 127 | PRKAG1 | 22135 | 18092 | -1.496 | -0.2283 | Yes | ||
| 128 | ZHX2 | 11842 | 18096 | -1.501 | -0.1945 | Yes | ||
| 129 | DOCK3 | 9920 5537 10751 | 18169 | -1.583 | -0.1626 | Yes | ||
| 130 | CHCHD7 | 16280 | 18243 | -1.684 | -0.1285 | Yes | ||
| 131 | TRPS1 | 8195 | 18344 | -1.859 | -0.0919 | Yes | ||
| 132 | GFRA1 | 9015 | 18358 | -1.890 | -0.0498 | Yes | ||
| 133 | SUPT4H1 | 9938 | 18579 | -2.818 | 0.0020 | Yes |

