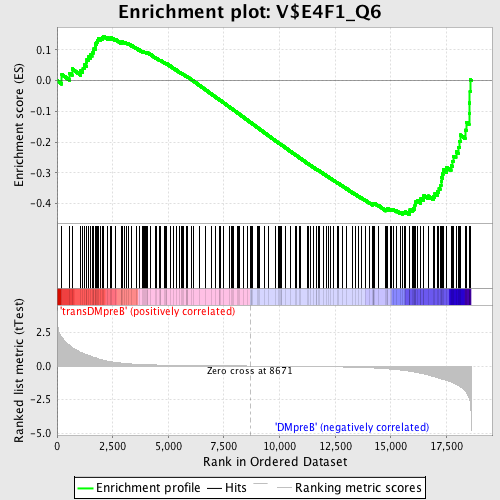
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | V$E4F1_Q6 |
| Enrichment Score (ES) | -0.43565184 |
| Normalized Enrichment Score (NES) | -1.2472403 |
| Nominal p-value | 0.051194537 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | USP48 | 16022 | 211 | 2.138 | 0.0200 | No | ||
| 2 | IKBKB | 4907 | 558 | 1.550 | 0.0239 | No | ||
| 3 | PURB | 5335 | 674 | 1.394 | 0.0382 | No | ||
| 4 | SRPR | 12525 7433 | 1064 | 1.010 | 0.0319 | No | ||
| 5 | GNAS | 9025 2963 2752 | 1162 | 0.961 | 0.0408 | No | ||
| 6 | PTMA | 9657 | 1221 | 0.916 | 0.0511 | No | ||
| 7 | RANBP2 | 20019 | 1330 | 0.846 | 0.0576 | No | ||
| 8 | HIC2 | 7185 | 1337 | 0.842 | 0.0697 | No | ||
| 9 | ATP2A2 | 4421 3481 | 1416 | 0.790 | 0.0770 | No | ||
| 10 | RAB6A | 18173 3905 | 1495 | 0.744 | 0.0837 | No | ||
| 11 | SNF1LK | 23033 | 1570 | 0.699 | 0.0900 | No | ||
| 12 | CBX8 | 11401 | 1617 | 0.672 | 0.0973 | No | ||
| 13 | OSR1 | 18971 | 1655 | 0.653 | 0.1049 | No | ||
| 14 | GOLGA5 | 21179 | 1714 | 0.620 | 0.1109 | No | ||
| 15 | EIF5A | 11345 20379 6590 | 1726 | 0.615 | 0.1193 | No | ||
| 16 | NEDD9 | 3206 21483 | 1777 | 0.580 | 0.1251 | No | ||
| 17 | SLC20A2 | 18654 | 1823 | 0.552 | 0.1308 | No | ||
| 18 | LENG9 | 6337 | 1838 | 0.542 | 0.1380 | No | ||
| 19 | DDX3X | 24379 | 1968 | 0.472 | 0.1379 | No | ||
| 20 | NEK1 | 18610 | 2042 | 0.441 | 0.1404 | No | ||
| 21 | CKB | 4523 | 2083 | 0.426 | 0.1445 | No | ||
| 22 | SMARCE1 | 1218 20253 | 2283 | 0.359 | 0.1390 | No | ||
| 23 | GRK6 | 6449 | 2385 | 0.327 | 0.1383 | No | ||
| 24 | DHX36 | 15328 | 2448 | 0.306 | 0.1394 | No | ||
| 25 | SLC18A2 | 5639 | 2635 | 0.262 | 0.1332 | No | ||
| 26 | SFRS1 | 8492 | 2888 | 0.219 | 0.1227 | No | ||
| 27 | EPC1 | 2039 8907 2033 | 2891 | 0.217 | 0.1258 | No | ||
| 28 | KCNA6 | 664 16990 665 4938 | 2931 | 0.210 | 0.1268 | No | ||
| 29 | CRY2 | 14518 | 3025 | 0.197 | 0.1246 | No | ||
| 30 | ALDOC | 20759 311 | 3129 | 0.182 | 0.1217 | No | ||
| 31 | PDE7A | 9544 1788 | 3212 | 0.170 | 0.1198 | No | ||
| 32 | NR6A1 | 14596 | 3360 | 0.151 | 0.1140 | No | ||
| 33 | RFX1 | 18548 | 3568 | 0.133 | 0.1047 | No | ||
| 34 | CD160 | 12023 | 3690 | 0.123 | 0.1000 | No | ||
| 35 | ARF4 | 22072 | 3830 | 0.112 | 0.0941 | No | ||
| 36 | CALM2 | 8681 | 3860 | 0.110 | 0.0941 | No | ||
| 37 | HABP4 | 12145 | 3944 | 0.105 | 0.0912 | No | ||
| 38 | CREB1 | 3990 8782 4558 4093 | 3961 | 0.105 | 0.0919 | No | ||
| 39 | NINJ1 | 21645 | 4006 | 0.102 | 0.0910 | No | ||
| 40 | NR3C1 | 9043 | 4051 | 0.099 | 0.0900 | No | ||
| 41 | DDX51 | 16752 12813 7636 | 4071 | 0.097 | 0.0904 | No | ||
| 42 | ERF | 4680 8914 | 4183 | 0.090 | 0.0857 | No | ||
| 43 | ATP10D | 16831 | 4431 | 0.079 | 0.0735 | No | ||
| 44 | GOLGA2 | 15039 2779 2923 | 4478 | 0.077 | 0.0721 | No | ||
| 45 | CD2AP | 22975 | 4610 | 0.072 | 0.0661 | No | ||
| 46 | CYR61 | 9159 15145 9311 5024 | 4642 | 0.071 | 0.0654 | No | ||
| 47 | TRPC1 | 2984 19039 | 4644 | 0.071 | 0.0664 | No | ||
| 48 | TBX20 | 19196 3044 3000 | 4846 | 0.064 | 0.0564 | No | ||
| 49 | RPL41 | 12611 | 4870 | 0.063 | 0.0561 | No | ||
| 50 | SHC3 | 21465 | 4896 | 0.062 | 0.0557 | No | ||
| 51 | MAFF | 22423 2243 | 4938 | 0.061 | 0.0543 | No | ||
| 52 | UBAP1 | 7388 | 5075 | 0.057 | 0.0478 | No | ||
| 53 | PNMA3 | 24312 | 5239 | 0.052 | 0.0397 | No | ||
| 54 | PITX2 | 15424 1878 | 5344 | 0.050 | 0.0348 | No | ||
| 55 | FGF14 | 21716 3005 3463 | 5520 | 0.046 | 0.0260 | No | ||
| 56 | RUSC1 | 1853 15283 | 5599 | 0.044 | 0.0224 | No | ||
| 57 | FLRT3 | 7730 12930 2673 | 5651 | 0.043 | 0.0203 | No | ||
| 58 | ATF1 | 8634 4417 | 5689 | 0.042 | 0.0189 | No | ||
| 59 | RFX5 | 7018 | 5801 | 0.040 | 0.0134 | No | ||
| 60 | EGR3 | 4656 | 5820 | 0.039 | 0.0130 | No | ||
| 61 | TAF10 | 10785 | 5865 | 0.038 | 0.0112 | No | ||
| 62 | NR4A1 | 9099 | 5872 | 0.038 | 0.0114 | No | ||
| 63 | C1QTNF7 | 16852 | 6028 | 0.035 | 0.0036 | No | ||
| 64 | GRM3 | 16929 | 6116 | 0.034 | -0.0007 | No | ||
| 65 | EGR2 | 8886 | 6380 | 0.030 | -0.0145 | No | ||
| 66 | NR4A3 | 9473 16212 5183 | 6648 | 0.025 | -0.0286 | No | ||
| 67 | IRX6 | 7225 | 6949 | 0.021 | -0.0446 | No | ||
| 68 | FOSB | 17945 | 7115 | 0.019 | -0.0532 | No | ||
| 69 | CHGB | 14835 | 7280 | 0.017 | -0.0619 | No | ||
| 70 | CALD1 | 4273 8463 | 7286 | 0.016 | -0.0619 | No | ||
| 71 | HOXB3 | 20685 | 7352 | 0.016 | -0.0652 | No | ||
| 72 | GNG4 | 21706 | 7491 | 0.014 | -0.0725 | No | ||
| 73 | HMGB1 | 9094 4855 | 7757 | 0.010 | -0.0867 | No | ||
| 74 | UBQLN2 | 896 24223 897 | 7829 | 0.009 | -0.0904 | No | ||
| 75 | BRUNOL6 | 19421 | 7898 | 0.009 | -0.0940 | No | ||
| 76 | DACT1 | 7203 | 7922 | 0.008 | -0.0951 | No | ||
| 77 | STAG1 | 5520 9905 2987 | 8108 | 0.006 | -0.1051 | No | ||
| 78 | FSTL5 | 10018 | 8168 | 0.006 | -0.1082 | No | ||
| 79 | HS6ST2 | 6939 2646 | 8207 | 0.005 | -0.1102 | No | ||
| 80 | SSTR2 | 20609 | 8377 | 0.003 | -0.1193 | No | ||
| 81 | ARG2 | 21228 | 8566 | 0.001 | -0.1295 | No | ||
| 82 | SATB2 | 5604 | 8702 | -0.000 | -0.1368 | No | ||
| 83 | PTP4A1 | 9658 | 8725 | -0.001 | -0.1380 | No | ||
| 84 | GRIN2B | 16957 | 8734 | -0.001 | -0.1384 | No | ||
| 85 | OSBP | 4278 | 8761 | -0.001 | -0.1398 | No | ||
| 86 | MASTL | 12471 7387 14674 | 8989 | -0.004 | -0.1520 | No | ||
| 87 | NKX2-2 | 5176 | 9059 | -0.005 | -0.1557 | No | ||
| 88 | SCAMP5 | 3028 19100 | 9089 | -0.005 | -0.1572 | No | ||
| 89 | ANK2 | 4275 | 9301 | -0.008 | -0.1685 | No | ||
| 90 | MRC2 | 1386 5117 | 9510 | -0.011 | -0.1797 | No | ||
| 91 | ZNF462 | 6321 | 9821 | -0.014 | -0.1963 | No | ||
| 92 | FOXA1 | 21060 | 9938 | -0.016 | -0.2023 | No | ||
| 93 | SYNGR3 | 23087 | 9995 | -0.017 | -0.2051 | No | ||
| 94 | PRR3 | 22995 | 10062 | -0.018 | -0.2084 | No | ||
| 95 | PKP4 | 2822 15008 | 10063 | -0.018 | -0.2082 | No | ||
| 96 | NUMBL | 1663 9496 | 10090 | -0.018 | -0.2093 | No | ||
| 97 | ATP6V0C | 8643 | 10268 | -0.020 | -0.2186 | No | ||
| 98 | SOX9 | 20611 | 10472 | -0.023 | -0.2293 | No | ||
| 99 | SACM1L | 19253 | 10708 | -0.027 | -0.2417 | No | ||
| 100 | MCAM | 3109 19479 | 10743 | -0.027 | -0.2431 | No | ||
| 101 | FKBP7 | 14554 | 10914 | -0.030 | -0.2519 | No | ||
| 102 | HNRPA2B1 | 1187 7001 7000 1040 | 10954 | -0.030 | -0.2536 | No | ||
| 103 | CLDN7 | 20817 | 11238 | -0.034 | -0.2684 | No | ||
| 104 | FOXD3 | 9088 | 11289 | -0.035 | -0.2706 | No | ||
| 105 | TRIB1 | 22467 | 11393 | -0.037 | -0.2757 | No | ||
| 106 | CALCRL | 12041 | 11508 | -0.039 | -0.2813 | No | ||
| 107 | PLK4 | 1870 1874 | 11649 | -0.042 | -0.2882 | No | ||
| 108 | KIRREL3 | 12571 | 11668 | -0.042 | -0.2886 | No | ||
| 109 | NAP1L2 | 24087 | 11755 | -0.044 | -0.2926 | No | ||
| 110 | SCG2 | 5410 14425 | 11801 | -0.045 | -0.2944 | No | ||
| 111 | ZBTB4 | 13267 | 11987 | -0.049 | -0.3037 | No | ||
| 112 | POLR2A | 5394 | 12113 | -0.052 | -0.3097 | No | ||
| 113 | CBX3 | 8705 4484 | 12195 | -0.054 | -0.3134 | No | ||
| 114 | GEM | 16274 | 12291 | -0.056 | -0.3177 | No | ||
| 115 | SLC36A3 | 20447 | 12424 | -0.060 | -0.3240 | No | ||
| 116 | NEUROD6 | 17138 | 12610 | -0.065 | -0.3330 | No | ||
| 117 | DNM3 | 5948 | 12637 | -0.066 | -0.3335 | No | ||
| 118 | MAP1A | 5124 | 12841 | -0.072 | -0.3434 | No | ||
| 119 | TLE3 | 5767 19416 | 13026 | -0.079 | -0.3523 | No | ||
| 120 | CXCL16 | 7251 | 13291 | -0.089 | -0.3653 | No | ||
| 121 | YWHAG | 16339 | 13409 | -0.094 | -0.3702 | No | ||
| 122 | KCNA2 | 15458 | 13547 | -0.101 | -0.3762 | No | ||
| 123 | MAP3K13 | 22814 | 13677 | -0.108 | -0.3816 | No | ||
| 124 | CCNI | 4493 | 13855 | -0.119 | -0.3895 | No | ||
| 125 | CLDN3 | 16680 | 14051 | -0.130 | -0.3981 | No | ||
| 126 | SMARCA1 | 24165 | 14183 | -0.140 | -0.4032 | No | ||
| 127 | PPARGC1A | 16533 | 14203 | -0.142 | -0.4021 | No | ||
| 128 | FGF9 | 8966 | 14217 | -0.143 | -0.4007 | No | ||
| 129 | KDELR3 | 2212 22422 | 14220 | -0.144 | -0.3987 | No | ||
| 130 | PPP2R2A | 3222 21774 | 14266 | -0.147 | -0.3990 | No | ||
| 131 | EGR4 | 24483 | 14440 | -0.164 | -0.4060 | No | ||
| 132 | LTBP1 | 1536 1604 6503 1559 1564 1518 | 14780 | -0.199 | -0.4214 | No | ||
| 133 | GTF2A1 | 2064 8187 8188 13512 | 14827 | -0.206 | -0.4209 | No | ||
| 134 | RHOBTB1 | 12789 | 14848 | -0.209 | -0.4189 | No | ||
| 135 | HDAC6 | 24197 | 14856 | -0.210 | -0.4162 | No | ||
| 136 | NOL4 | 2028 11432 | 14973 | -0.225 | -0.4192 | No | ||
| 137 | FZD7 | 14242 | 15014 | -0.230 | -0.4180 | No | ||
| 138 | EIF4A1 | 8889 23719 | 15101 | -0.240 | -0.4192 | No | ||
| 139 | MYST1 | 18059 | 15257 | -0.264 | -0.4237 | No | ||
| 140 | HIST1H4H | 7618 | 15428 | -0.294 | -0.4286 | No | ||
| 141 | HOXC10 | 22342 | 15539 | -0.316 | -0.4299 | No | ||
| 142 | SLC31A1 | 2447 16195 | 15611 | -0.334 | -0.4289 | No | ||
| 143 | PSMD12 | 20621 | 15664 | -0.347 | -0.4266 | No | ||
| 144 | RPS6KA3 | 8490 | 15832 | -0.385 | -0.4300 | Yes | ||
| 145 | GPR3 | 15729 | 15836 | -0.386 | -0.4245 | Yes | ||
| 146 | EDG5 | 19216 | 15859 | -0.392 | -0.4199 | Yes | ||
| 147 | AGPAT1 | 23275 | 15995 | -0.430 | -0.4210 | Yes | ||
| 148 | PSME3 | 20657 | 16006 | -0.433 | -0.4151 | Yes | ||
| 149 | PDLIM3 | 18625 | 16078 | -0.452 | -0.4124 | Yes | ||
| 150 | ZADH2 | 23501 | 16081 | -0.453 | -0.4058 | Yes | ||
| 151 | EHD4 | 8275 | 16090 | -0.455 | -0.3996 | Yes | ||
| 152 | PPM2C | 491 | 16124 | -0.464 | -0.3945 | Yes | ||
| 153 | SLC25A25 | 10431 | 16194 | -0.485 | -0.3912 | Yes | ||
| 154 | ELOVL5 | 19373 | 16337 | -0.532 | -0.3911 | Yes | ||
| 155 | TBRG4 | 1422 20529 | 16349 | -0.535 | -0.3838 | Yes | ||
| 156 | ZNF24 | 7207 | 16461 | -0.573 | -0.3814 | Yes | ||
| 157 | PTOV1 | 13522 | 16483 | -0.581 | -0.3740 | Yes | ||
| 158 | NDUFB2 | 7542 | 16689 | -0.666 | -0.3754 | Yes | ||
| 159 | NR4A2 | 5203 | 16920 | -0.781 | -0.3764 | Yes | ||
| 160 | BCL2L13 | 17302 | 16980 | -0.816 | -0.3676 | Yes | ||
| 161 | SEC63 | 8943 4708 | 17100 | -0.878 | -0.3611 | Yes | ||
| 162 | PCQAP | 1661 13581 | 17164 | -0.903 | -0.3513 | Yes | ||
| 163 | ETF1 | 23467 | 17215 | -0.931 | -0.3403 | Yes | ||
| 164 | ABCF3 | 6578 | 17260 | -0.955 | -0.3287 | Yes | ||
| 165 | MORF4L2 | 12118 | 17275 | -0.963 | -0.3153 | Yes | ||
| 166 | FAU | 8954 | 17327 | -0.991 | -0.3035 | Yes | ||
| 167 | PAFAH1B1 | 1340 5220 9524 | 17349 | -0.998 | -0.2900 | Yes | ||
| 168 | CSTF3 | 10449 6002 | 17511 | -1.069 | -0.2831 | Yes | ||
| 169 | SH2D2A | 6569 | 17707 | -1.183 | -0.2763 | Yes | ||
| 170 | WNT10A | 14221 | 17775 | -1.232 | -0.2618 | Yes | ||
| 171 | SLC30A5 | 3190 21361 | 17818 | -1.266 | -0.2455 | Yes | ||
| 172 | TNFAIP8 | 8391 | 17930 | -1.352 | -0.2317 | Yes | ||
| 173 | RNF5 | 63 | 18034 | -1.434 | -0.2162 | Yes | ||
| 174 | PRKAG1 | 22135 | 18092 | -1.496 | -0.1973 | Yes | ||
| 175 | DUSP1 | 23061 | 18133 | -1.547 | -0.1768 | Yes | ||
| 176 | PPP6C | 7491 | 18340 | -1.848 | -0.1608 | Yes | ||
| 177 | CCBL1 | 14630 | 18389 | -1.931 | -0.1351 | Yes | ||
| 178 | RAB1A | 20942 | 18517 | -2.350 | -0.1075 | Yes | ||
| 179 | MAP1LC3A | 14774 2741 | 18529 | -2.408 | -0.0727 | Yes | ||
| 180 | WBP5 | 10297 | 18557 | -2.578 | -0.0363 | Yes | ||
| 181 | MRPL49 | 3696 9472 23990 | 18568 | -2.689 | 0.0026 | Yes |

