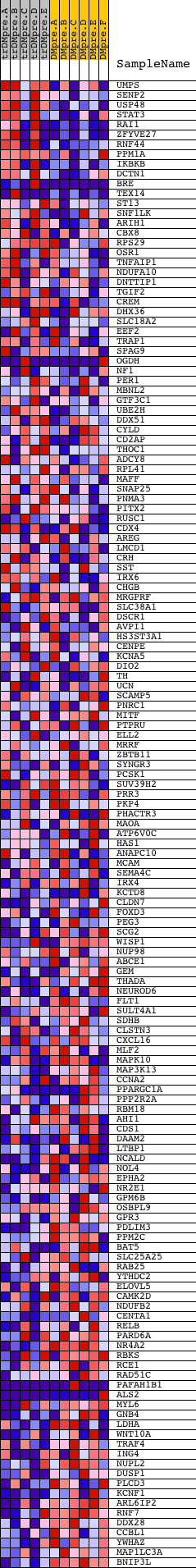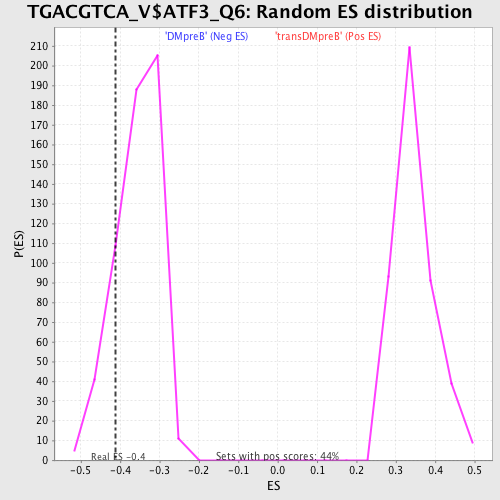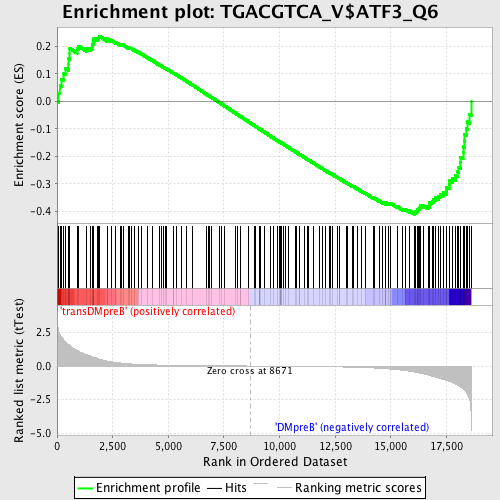
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | TGACGTCA_V$ATF3_Q6 |
| Enrichment Score (ES) | -0.41177195 |
| Normalized Enrichment Score (NES) | -1.159059 |
| Nominal p-value | 0.15026833 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | UMPS | 22606 1760 | 83 | 2.546 | 0.0290 | No | ||
| 2 | SENP2 | 7990 | 129 | 2.334 | 0.0572 | No | ||
| 3 | USP48 | 16022 | 211 | 2.138 | 0.0810 | No | ||
| 4 | STAT3 | 5525 9906 | 302 | 1.915 | 0.1013 | No | ||
| 5 | RAI1 | 5355 20861 | 394 | 1.778 | 0.1197 | No | ||
| 6 | ZFYVE27 | 23849 | 527 | 1.593 | 0.1335 | No | ||
| 7 | RNF44 | 8364 | 529 | 1.588 | 0.1544 | No | ||
| 8 | PPM1A | 5279 9607 | 556 | 1.551 | 0.1734 | No | ||
| 9 | IKBKB | 4907 | 558 | 1.550 | 0.1937 | No | ||
| 10 | DCTN1 | 1100 17392 | 895 | 1.166 | 0.1908 | No | ||
| 11 | BRE | 3640 4226 16879 | 974 | 1.078 | 0.2008 | No | ||
| 12 | TEX14 | 20714 | 1313 | 0.852 | 0.1937 | No | ||
| 13 | ST13 | 12865 | 1500 | 0.738 | 0.1933 | No | ||
| 14 | SNF1LK | 23033 | 1570 | 0.699 | 0.1988 | No | ||
| 15 | ARIH1 | 6210 | 1581 | 0.693 | 0.2073 | No | ||
| 16 | CBX8 | 11401 | 1617 | 0.672 | 0.2143 | No | ||
| 17 | RPS29 | 9754 | 1647 | 0.656 | 0.2213 | No | ||
| 18 | OSR1 | 18971 | 1655 | 0.653 | 0.2295 | No | ||
| 19 | TNFAIP1 | 20333 | 1802 | 0.561 | 0.2290 | No | ||
| 20 | NDUFA10 | 7420 | 1880 | 0.518 | 0.2316 | No | ||
| 21 | DNTTIP1 | 14736 | 1905 | 0.505 | 0.2370 | No | ||
| 22 | TGIF2 | 6013 6012 | 2245 | 0.371 | 0.2235 | No | ||
| 23 | CREM | 2005 1965 23496 1951 | 2274 | 0.362 | 0.2268 | No | ||
| 24 | DHX36 | 15328 | 2448 | 0.306 | 0.2214 | No | ||
| 25 | SLC18A2 | 5639 | 2635 | 0.262 | 0.2148 | No | ||
| 26 | EEF2 | 8881 4654 8882 | 2867 | 0.222 | 0.2052 | No | ||
| 27 | TRAP1 | 22683 | 2911 | 0.213 | 0.2057 | No | ||
| 28 | SPAG9 | 1322 12904 7707 | 2979 | 0.203 | 0.2047 | No | ||
| 29 | OGDH | 9502 1412 | 3215 | 0.170 | 0.1942 | No | ||
| 30 | NF1 | 5165 | 3253 | 0.164 | 0.1944 | No | ||
| 31 | PER1 | 20827 | 3332 | 0.155 | 0.1922 | No | ||
| 32 | MBNL2 | 21932 | 3499 | 0.138 | 0.1850 | No | ||
| 33 | GTF3C1 | 10607 17646 | 3650 | 0.126 | 0.1785 | No | ||
| 34 | UBE2H | 5823 5822 | 3809 | 0.114 | 0.1715 | No | ||
| 35 | DDX51 | 16752 12813 7636 | 4071 | 0.097 | 0.1586 | No | ||
| 36 | CYLD | 18532 | 4266 | 0.086 | 0.1492 | No | ||
| 37 | CD2AP | 22975 | 4610 | 0.072 | 0.1316 | No | ||
| 38 | THOC1 | 23623 | 4686 | 0.069 | 0.1285 | No | ||
| 39 | ADCY8 | 22281 | 4797 | 0.065 | 0.1234 | No | ||
| 40 | RPL41 | 12611 | 4870 | 0.063 | 0.1203 | No | ||
| 41 | MAFF | 22423 2243 | 4938 | 0.061 | 0.1175 | No | ||
| 42 | SNAP25 | 5466 | 5234 | 0.052 | 0.1022 | No | ||
| 43 | PNMA3 | 24312 | 5239 | 0.052 | 0.1026 | No | ||
| 44 | PITX2 | 15424 1878 | 5344 | 0.050 | 0.0977 | No | ||
| 45 | RUSC1 | 1853 15283 | 5599 | 0.044 | 0.0845 | No | ||
| 46 | CDX4 | 24273 | 5828 | 0.039 | 0.0727 | No | ||
| 47 | AREG | 16796 | 6082 | 0.034 | 0.0594 | No | ||
| 48 | LMCD1 | 17337 1117 | 6705 | 0.024 | 0.0260 | No | ||
| 49 | CRH | 8784 | 6825 | 0.023 | 0.0199 | No | ||
| 50 | SST | 22625 | 6831 | 0.023 | 0.0199 | No | ||
| 51 | IRX6 | 7225 | 6949 | 0.021 | 0.0139 | No | ||
| 52 | CHGB | 14835 | 7280 | 0.017 | -0.0038 | No | ||
| 53 | MRGPRF | 17995 | 7387 | 0.015 | -0.0093 | No | ||
| 54 | SLC38A1 | 22148 | 7517 | 0.013 | -0.0161 | No | ||
| 55 | DSCR1 | 22536 1633 | 8007 | 0.008 | -0.0425 | No | ||
| 56 | AVPI1 | 23675 | 8097 | 0.007 | -0.0473 | No | ||
| 57 | HS3ST3A1 | 20844 | 8223 | 0.005 | -0.0540 | No | ||
| 58 | CENPE | 15414 | 8244 | 0.005 | -0.0550 | No | ||
| 59 | KCNA5 | 9203 | 8590 | 0.001 | -0.0736 | No | ||
| 60 | DIO2 | 21014 2159 | 8884 | -0.002 | -0.0895 | No | ||
| 61 | TH | 17548 | 8917 | -0.003 | -0.0912 | No | ||
| 62 | UCN | 16573 | 9083 | -0.005 | -0.1001 | No | ||
| 63 | SCAMP5 | 3028 19100 | 9089 | -0.005 | -0.1003 | No | ||
| 64 | PNRC1 | 15925 | 9096 | -0.005 | -0.1005 | No | ||
| 65 | MITF | 17349 | 9128 | -0.005 | -0.1021 | No | ||
| 66 | PTPRU | 15738 | 9303 | -0.008 | -0.1114 | No | ||
| 67 | ELL2 | 5329 | 9594 | -0.012 | -0.1270 | No | ||
| 68 | MRRF | 12594 2837 | 9709 | -0.013 | -0.1330 | No | ||
| 69 | ZBTB11 | 11309 | 9904 | -0.016 | -0.1433 | No | ||
| 70 | SYNGR3 | 23087 | 9995 | -0.017 | -0.1480 | No | ||
| 71 | PCSK1 | 21600 | 10056 | -0.018 | -0.1510 | No | ||
| 72 | SUV39H2 | 7232 12263 | 10059 | -0.018 | -0.1509 | No | ||
| 73 | PRR3 | 22995 | 10062 | -0.018 | -0.1507 | No | ||
| 74 | PKP4 | 2822 15008 | 10063 | -0.018 | -0.1505 | No | ||
| 75 | PHACTR3 | 7900 2770 | 10093 | -0.018 | -0.1518 | No | ||
| 76 | MAOA | 9368 24374 | 10188 | -0.019 | -0.1567 | No | ||
| 77 | ATP6V0C | 8643 | 10268 | -0.020 | -0.1607 | No | ||
| 78 | HAS1 | 23116 | 10401 | -0.022 | -0.1675 | No | ||
| 79 | ANAPC10 | 7595 12760 7596 | 10718 | -0.027 | -0.1843 | No | ||
| 80 | MCAM | 3109 19479 | 10743 | -0.027 | -0.1852 | No | ||
| 81 | SEMA4C | 9799 | 10878 | -0.029 | -0.1921 | No | ||
| 82 | IRX4 | 11944 | 10881 | -0.029 | -0.1918 | No | ||
| 83 | KCTD8 | 16523 | 11138 | -0.033 | -0.2053 | No | ||
| 84 | CLDN7 | 20817 | 11238 | -0.034 | -0.2102 | No | ||
| 85 | FOXD3 | 9088 | 11289 | -0.035 | -0.2124 | No | ||
| 86 | PEG3 | 5240 | 11533 | -0.039 | -0.2251 | No | ||
| 87 | SCG2 | 5410 14425 | 11801 | -0.045 | -0.2389 | No | ||
| 88 | WISP1 | 22463 2289 | 11932 | -0.047 | -0.2453 | No | ||
| 89 | NUP98 | 17726 | 12065 | -0.050 | -0.2518 | No | ||
| 90 | ABCE1 | 6270 | 12231 | -0.055 | -0.2600 | No | ||
| 91 | GEM | 16274 | 12291 | -0.056 | -0.2625 | No | ||
| 92 | THADA | 11614 | 12359 | -0.058 | -0.2654 | No | ||
| 93 | NEUROD6 | 17138 | 12610 | -0.065 | -0.2781 | No | ||
| 94 | FLT1 | 3483 16287 | 12672 | -0.067 | -0.2805 | No | ||
| 95 | SULT4A1 | 22179 | 12681 | -0.067 | -0.2800 | No | ||
| 96 | SDHB | 2348 12566 14880 | 12985 | -0.078 | -0.2954 | No | ||
| 97 | CLSTN3 | 17006 | 13070 | -0.080 | -0.2989 | No | ||
| 98 | CXCL16 | 7251 | 13291 | -0.089 | -0.3097 | No | ||
| 99 | MLF2 | 17283 | 13321 | -0.090 | -0.3100 | No | ||
| 100 | MAPK10 | 11169 | 13511 | -0.099 | -0.3190 | No | ||
| 101 | MAP3K13 | 22814 | 13677 | -0.108 | -0.3265 | No | ||
| 102 | CCNA2 | 15357 | 13871 | -0.120 | -0.3353 | No | ||
| 103 | PPARGC1A | 16533 | 14203 | -0.142 | -0.3514 | No | ||
| 104 | PPP2R2A | 3222 21774 | 14266 | -0.147 | -0.3528 | No | ||
| 105 | RBM18 | 2789 14604 | 14483 | -0.168 | -0.3623 | No | ||
| 106 | AHI1 | 20077 | 14640 | -0.184 | -0.3684 | No | ||
| 107 | CDS1 | 16779 | 14748 | -0.196 | -0.3716 | No | ||
| 108 | DAAM2 | 22945 176 | 14770 | -0.198 | -0.3701 | No | ||
| 109 | LTBP1 | 1536 1604 6503 1559 1564 1518 | 14780 | -0.199 | -0.3680 | No | ||
| 110 | NCALD | 22310 | 14916 | -0.218 | -0.3724 | No | ||
| 111 | NOL4 | 2028 11432 | 14973 | -0.225 | -0.3725 | No | ||
| 112 | EPHA2 | 16006 | 14984 | -0.226 | -0.3701 | No | ||
| 113 | NR2E1 | 19780 | 15278 | -0.269 | -0.3824 | No | ||
| 114 | GPM6B | 9034 | 15537 | -0.316 | -0.3922 | No | ||
| 115 | OSBPL9 | 4141 | 15654 | -0.343 | -0.3940 | No | ||
| 116 | GPR3 | 15729 | 15836 | -0.386 | -0.3987 | No | ||
| 117 | PDLIM3 | 18625 | 16078 | -0.452 | -0.4058 | Yes | ||
| 118 | PPM2C | 491 | 16124 | -0.464 | -0.4022 | Yes | ||
| 119 | BAT5 | 23261 | 16177 | -0.480 | -0.3987 | Yes | ||
| 120 | SLC25A25 | 10431 | 16194 | -0.485 | -0.3932 | Yes | ||
| 121 | RAB25 | 15287 | 16264 | -0.502 | -0.3903 | Yes | ||
| 122 | YTHDC2 | 23564 | 16309 | -0.522 | -0.3858 | Yes | ||
| 123 | ELOVL5 | 19373 | 16337 | -0.532 | -0.3803 | Yes | ||
| 124 | CAMK2D | 4232 | 16448 | -0.567 | -0.3788 | Yes | ||
| 125 | NDUFB2 | 7542 | 16689 | -0.666 | -0.3830 | Yes | ||
| 126 | CENTA1 | 10546 16319 | 16732 | -0.690 | -0.3762 | Yes | ||
| 127 | RELB | 17942 | 16740 | -0.696 | -0.3675 | Yes | ||
| 128 | PARD6A | 12142 | 16854 | -0.748 | -0.3637 | Yes | ||
| 129 | NR4A2 | 5203 | 16920 | -0.781 | -0.3570 | Yes | ||
| 130 | RBKS | 16568 | 17022 | -0.840 | -0.3514 | Yes | ||
| 131 | RCE1 | 23965 | 17159 | -0.902 | -0.3469 | Yes | ||
| 132 | RAD51C | 4330 | 17248 | -0.948 | -0.3392 | Yes | ||
| 133 | PAFAH1B1 | 1340 5220 9524 | 17349 | -0.998 | -0.3315 | Yes | ||
| 134 | ALS2 | 7882 13151 | 17484 | -1.055 | -0.3249 | Yes | ||
| 135 | MYL6 | 9438 3408 | 17517 | -1.071 | -0.3125 | Yes | ||
| 136 | GNB4 | 4787 9028 | 17633 | -1.139 | -0.3038 | Yes | ||
| 137 | LDHA | 9269 | 17646 | -1.146 | -0.2893 | Yes | ||
| 138 | WNT10A | 14221 | 17775 | -1.232 | -0.2801 | Yes | ||
| 139 | TRAF4 | 10217 5796 1400 | 17912 | -1.330 | -0.2700 | Yes | ||
| 140 | ING4 | 6602 1155 1102 11371 | 17993 | -1.400 | -0.2559 | Yes | ||
| 141 | NUPL2 | 6072 | 18061 | -1.465 | -0.2402 | Yes | ||
| 142 | DUSP1 | 23061 | 18133 | -1.547 | -0.2237 | Yes | ||
| 143 | PLCD3 | 20200 | 18141 | -1.559 | -0.2036 | Yes | ||
| 144 | KCNF1 | 19140 | 18246 | -1.688 | -0.1870 | Yes | ||
| 145 | ARL6IP2 | 1496 7090 12093 | 18252 | -1.701 | -0.1650 | Yes | ||
| 146 | RNF7 | 19036 | 18307 | -1.790 | -0.1443 | Yes | ||
| 147 | DDX28 | 18758 | 18325 | -1.829 | -0.1212 | Yes | ||
| 148 | CCBL1 | 14630 | 18389 | -1.931 | -0.0992 | Yes | ||
| 149 | YWHAZ | 10370 | 18444 | -2.073 | -0.0749 | Yes | ||
| 150 | MAP1LC3A | 14774 2741 | 18529 | -2.408 | -0.0478 | Yes | ||
| 151 | BNIP3L | 21775 | 18614 | -3.987 | 0.0001 | Yes |