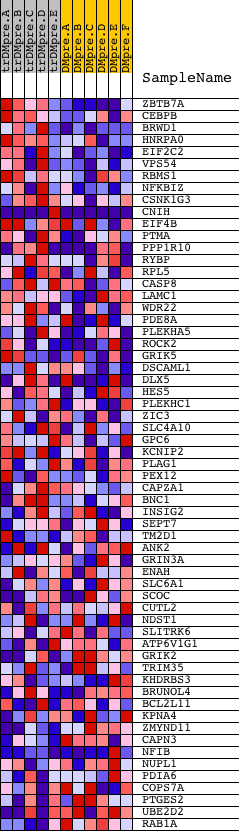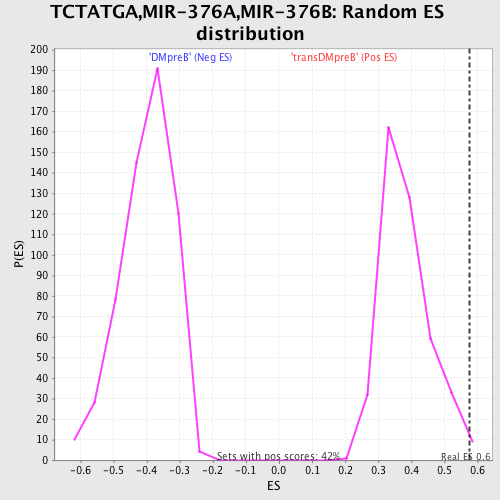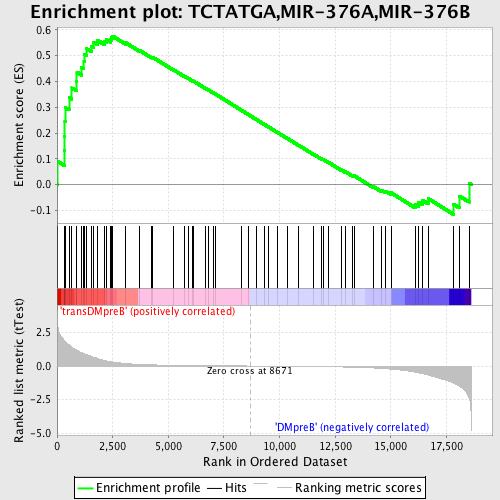
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | TCTATGA,MIR-376A,MIR-376B |
| Enrichment Score (ES) | 0.5754968 |
| Normalized Enrichment Score (NES) | 1.4995005 |
| Nominal p-value | 0.011792453 |
| FDR q-value | 0.17254807 |
| FWER p-Value | 0.836 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ZBTB7A | 5004 | 24 | 2.966 | 0.0904 | Yes | ||
| 2 | CEBPB | 8733 | 335 | 1.856 | 0.1311 | Yes | ||
| 3 | BRWD1 | 8222 1659 22532 | 342 | 1.842 | 0.1877 | Yes | ||
| 4 | HNRPA0 | 13382 | 351 | 1.829 | 0.2438 | Yes | ||
| 5 | EIF2C2 | 6247 | 370 | 1.803 | 0.2986 | Yes | ||
| 6 | VPS54 | 20941 | 539 | 1.568 | 0.3380 | Yes | ||
| 7 | RBMS1 | 14580 | 667 | 1.405 | 0.3746 | Yes | ||
| 8 | NFKBIZ | 22575 | 861 | 1.195 | 0.4011 | Yes | ||
| 9 | CSNK1G3 | 12873 | 892 | 1.169 | 0.4356 | Yes | ||
| 10 | CNIH | 4536 23971 | 1102 | 0.998 | 0.4552 | Yes | ||
| 11 | EIF4B | 13279 7979 | 1207 | 0.924 | 0.4782 | Yes | ||
| 12 | PTMA | 9657 | 1221 | 0.916 | 0.5058 | Yes | ||
| 13 | PPP1R10 | 6972 11960 6971 | 1316 | 0.850 | 0.5270 | Yes | ||
| 14 | RYBP | 17056 | 1553 | 0.704 | 0.5361 | Yes | ||
| 15 | RPL5 | 9746 | 1643 | 0.659 | 0.5517 | Yes | ||
| 16 | CASP8 | 8694 | 1829 | 0.549 | 0.5587 | Yes | ||
| 17 | LAMC1 | 13805 | 2145 | 0.401 | 0.5541 | Yes | ||
| 18 | WDR22 | 6803 | 2232 | 0.375 | 0.5610 | Yes | ||
| 19 | PDE8A | 18202 | 2417 | 0.317 | 0.5609 | Yes | ||
| 20 | PLEKHA5 | 8453 | 2435 | 0.310 | 0.5696 | Yes | ||
| 21 | ROCK2 | 21309 5387 | 2496 | 0.295 | 0.5755 | Yes | ||
| 22 | GRIK5 | 350 17930 | 3079 | 0.190 | 0.5500 | No | ||
| 23 | DSCAML1 | 4338 13224 | 3687 | 0.123 | 0.5211 | No | ||
| 24 | DLX5 | 17219 1058 | 4253 | 0.087 | 0.4933 | No | ||
| 25 | HES5 | 15973 | 4294 | 0.085 | 0.4938 | No | ||
| 26 | PLEKHC1 | 21864 | 5237 | 0.052 | 0.4446 | No | ||
| 27 | ZIC3 | 10430 | 5735 | 0.041 | 0.4191 | No | ||
| 28 | SLC4A10 | 15004 | 5885 | 0.038 | 0.4123 | No | ||
| 29 | GPC6 | 21937 | 6100 | 0.034 | 0.4018 | No | ||
| 30 | KCNIP2 | 23655 3725 | 6122 | 0.034 | 0.4017 | No | ||
| 31 | PLAG1 | 7148 15949 12161 | 6686 | 0.025 | 0.3721 | No | ||
| 32 | PEX12 | 20321 | 6785 | 0.023 | 0.3676 | No | ||
| 33 | CAPZA1 | 8687 | 7036 | 0.020 | 0.3547 | No | ||
| 34 | BNC1 | 17772 | 7132 | 0.018 | 0.3501 | No | ||
| 35 | INSIG2 | 3944 4124 4073 | 8293 | 0.004 | 0.2878 | No | ||
| 36 | SEPT7 | 10640 | 8617 | 0.001 | 0.2704 | No | ||
| 37 | TM2D1 | 13570 | 8941 | -0.003 | 0.2531 | No | ||
| 38 | ANK2 | 4275 | 9301 | -0.008 | 0.2340 | No | ||
| 39 | GRIN3A | 15883 | 9490 | -0.010 | 0.2241 | No | ||
| 40 | ENAH | 4665 8901 | 9897 | -0.015 | 0.2027 | No | ||
| 41 | SLC6A1 | 10557 | 10351 | -0.021 | 0.1790 | No | ||
| 42 | SCOC | 3821 12106 | 10840 | -0.028 | 0.1536 | No | ||
| 43 | CUTL2 | 13110 4578 | 11545 | -0.040 | 0.1169 | No | ||
| 44 | NDST1 | 23430 4885 | 11878 | -0.046 | 0.1004 | No | ||
| 45 | SLITRK6 | 21724 | 11958 | -0.048 | 0.0976 | No | ||
| 46 | ATP6V1G1 | 12322 | 12176 | -0.053 | 0.0876 | No | ||
| 47 | GRIK2 | 3318 | 12784 | -0.071 | 0.0570 | No | ||
| 48 | TRIM35 | 7348 | 12957 | -0.077 | 0.0501 | No | ||
| 49 | KHDRBS3 | 22462 | 13294 | -0.089 | 0.0348 | No | ||
| 50 | BRUNOL4 | 2010 4229 | 13352 | -0.091 | 0.0346 | No | ||
| 51 | BCL2L11 | 2790 14861 | 14199 | -0.141 | -0.0067 | No | ||
| 52 | KPNA4 | 15321 | 14580 | -0.177 | -0.0217 | No | ||
| 53 | ZMYND11 | 12364 3269 3286 3223 | 14778 | -0.199 | -0.0261 | No | ||
| 54 | CAPN3 | 8686 2868 | 15012 | -0.230 | -0.0316 | No | ||
| 55 | NFIB | 15855 | 16096 | -0.456 | -0.0759 | No | ||
| 56 | NUPL1 | 12967 7767 | 16250 | -0.498 | -0.0687 | No | ||
| 57 | PDIA6 | 21308 | 16435 | -0.562 | -0.0613 | No | ||
| 58 | COPS7A | 11221 16997 | 16680 | -0.662 | -0.0539 | No | ||
| 59 | PTGES2 | 15038 | 17825 | -1.270 | -0.0763 | No | ||
| 60 | UBE2D2 | 12148 7139 | 18087 | -1.494 | -0.0442 | No | ||
| 61 | RAB1A | 20942 | 18517 | -2.350 | 0.0053 | No |