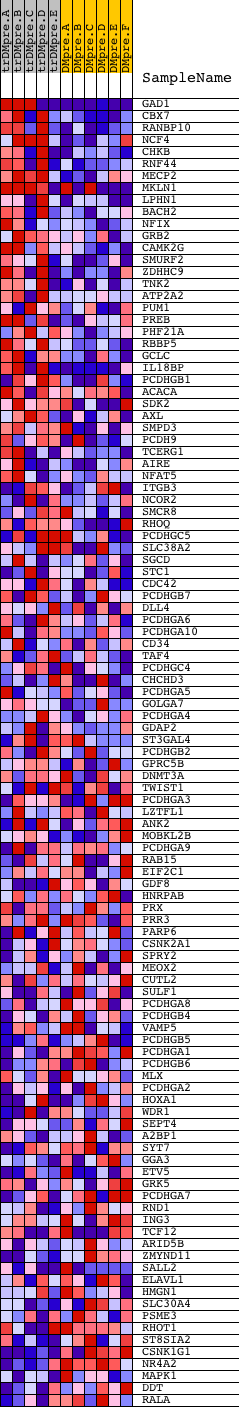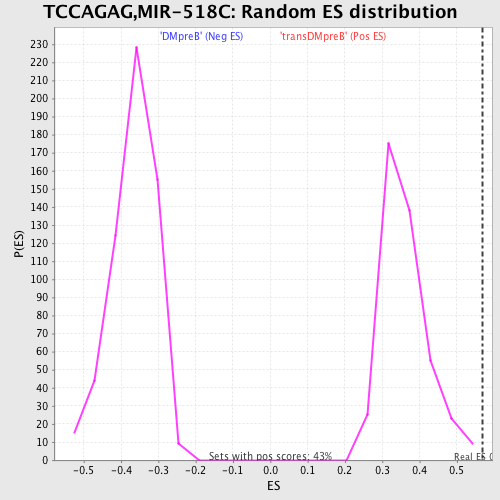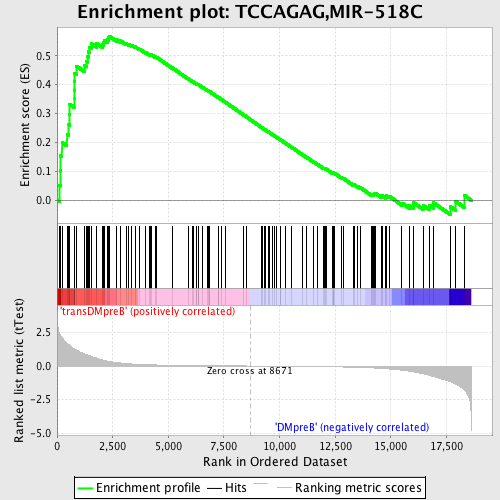
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | TCCAGAG,MIR-518C |
| Enrichment Score (ES) | 0.5678569 |
| Normalized Enrichment Score (NES) | 1.5753778 |
| Nominal p-value | 0.002352941 |
| FDR q-value | 0.20982109 |
| FWER p-Value | 0.477 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GAD1 | 14993 2690 | 104 | 2.429 | 0.0515 | Yes | ||
| 2 | CBX7 | 11973 | 142 | 2.265 | 0.1027 | Yes | ||
| 3 | RANBP10 | 18764 | 167 | 2.217 | 0.1535 | Yes | ||
| 4 | NCF4 | 22435 | 219 | 2.123 | 0.2006 | Yes | ||
| 5 | CHKB | 4518 | 444 | 1.707 | 0.2286 | Yes | ||
| 6 | RNF44 | 8364 | 529 | 1.588 | 0.2614 | Yes | ||
| 7 | MECP2 | 5088 | 557 | 1.551 | 0.2964 | Yes | ||
| 8 | MKLN1 | 6584 11339 | 568 | 1.535 | 0.3319 | Yes | ||
| 9 | LPHN1 | 6850 174 | 765 | 1.288 | 0.3516 | Yes | ||
| 10 | BACH2 | 8648 | 769 | 1.283 | 0.3816 | Yes | ||
| 11 | NFIX | 9458 | 780 | 1.276 | 0.4110 | Yes | ||
| 12 | GRB2 | 20149 | 792 | 1.266 | 0.4402 | Yes | ||
| 13 | CAMK2G | 21905 | 866 | 1.191 | 0.4642 | Yes | ||
| 14 | SMURF2 | 185 | 1242 | 0.903 | 0.4652 | Yes | ||
| 15 | ZDHHC9 | 24163 | 1320 | 0.849 | 0.4810 | Yes | ||
| 16 | TNK2 | 22784 | 1375 | 0.816 | 0.4972 | Yes | ||
| 17 | ATP2A2 | 4421 3481 | 1416 | 0.790 | 0.5136 | Yes | ||
| 18 | PUM1 | 8160 | 1456 | 0.766 | 0.5295 | Yes | ||
| 19 | PREB | 6948 3628 | 1542 | 0.714 | 0.5417 | Yes | ||
| 20 | PHF21A | 14943 | 1774 | 0.587 | 0.5430 | Yes | ||
| 21 | RBBP5 | 5620 5619 | 2056 | 0.437 | 0.5381 | Yes | ||
| 22 | GCLC | 19374 | 2099 | 0.420 | 0.5457 | Yes | ||
| 23 | IL18BP | 17730 | 2129 | 0.408 | 0.5537 | Yes | ||
| 24 | PCDHGB1 | 13532 8207 | 2266 | 0.363 | 0.5549 | Yes | ||
| 25 | ACACA | 309 4209 | 2298 | 0.356 | 0.5616 | Yes | ||
| 26 | SDK2 | 6208 20162 | 2333 | 0.344 | 0.5679 | Yes | ||
| 27 | AXL | 17922 | 2667 | 0.256 | 0.5559 | No | ||
| 28 | SMPD3 | 18757 | 2848 | 0.226 | 0.5515 | No | ||
| 29 | PCDH9 | 21736 | 3111 | 0.185 | 0.5417 | No | ||
| 30 | TCERG1 | 12075 2027 7068 | 3206 | 0.171 | 0.5406 | No | ||
| 31 | AIRE | 19716 3386 | 3357 | 0.152 | 0.5360 | No | ||
| 32 | NFAT5 | 3921 7037 12036 | 3519 | 0.137 | 0.5306 | No | ||
| 33 | ITGB3 | 20631 | 3713 | 0.121 | 0.5230 | No | ||
| 34 | NCOR2 | 9835 16367 | 3976 | 0.103 | 0.5112 | No | ||
| 35 | SMCR8 | 53 | 4168 | 0.091 | 0.5031 | No | ||
| 36 | RHOQ | 23142 | 4214 | 0.089 | 0.5027 | No | ||
| 37 | PCDHGC5 | 13540 8208 | 4247 | 0.087 | 0.5030 | No | ||
| 38 | SLC38A2 | 22149 2219 | 4413 | 0.080 | 0.4960 | No | ||
| 39 | SGCD | 1425 10779 | 4459 | 0.078 | 0.4954 | No | ||
| 40 | STC1 | 21974 | 5167 | 0.054 | 0.4585 | No | ||
| 41 | CDC42 | 4503 8722 4504 2465 | 5895 | 0.038 | 0.4201 | No | ||
| 42 | PCDHGB7 | 13537 | 6066 | 0.035 | 0.4117 | No | ||
| 43 | DLL4 | 7040 | 6145 | 0.034 | 0.4083 | No | ||
| 44 | PCDHGA6 | 13546 | 6245 | 0.032 | 0.4037 | No | ||
| 45 | PCDHGA10 | 13550 | 6286 | 0.031 | 0.4022 | No | ||
| 46 | CD34 | 14006 | 6346 | 0.030 | 0.3998 | No | ||
| 47 | TAF4 | 14319 | 6357 | 0.030 | 0.3999 | No | ||
| 48 | PCDHGC4 | 13539 | 6530 | 0.027 | 0.3913 | No | ||
| 49 | CHCHD3 | 17188 | 6773 | 0.023 | 0.3787 | No | ||
| 50 | PCDHGA5 | 13545 | 6804 | 0.023 | 0.3777 | No | ||
| 51 | GOLGA7 | 12188 | 6827 | 0.023 | 0.3770 | No | ||
| 52 | PCDHGA4 | 13544 | 6863 | 0.022 | 0.3756 | No | ||
| 53 | GDAP2 | 15481 | 7246 | 0.017 | 0.3554 | No | ||
| 54 | ST3GAL4 | 9816 | 7267 | 0.017 | 0.3547 | No | ||
| 55 | PCDHGB2 | 13533 | 7400 | 0.015 | 0.3479 | No | ||
| 56 | GPRC5B | 17662 | 7589 | 0.013 | 0.3381 | No | ||
| 57 | DNMT3A | 2167 21330 | 8385 | 0.003 | 0.2952 | No | ||
| 58 | TWIST1 | 21291 | 8501 | 0.002 | 0.2890 | No | ||
| 59 | PCDHGA3 | 13543 | 9202 | -0.006 | 0.2514 | No | ||
| 60 | LZTFL1 | 8210 | 9237 | -0.007 | 0.2497 | No | ||
| 61 | ANK2 | 4275 | 9301 | -0.008 | 0.2464 | No | ||
| 62 | MOBKL2B | 15922 | 9344 | -0.008 | 0.2444 | No | ||
| 63 | PCDHGA9 | 13549 | 9514 | -0.011 | 0.2355 | No | ||
| 64 | RAB15 | 21035 | 9564 | -0.011 | 0.2331 | No | ||
| 65 | EIF2C1 | 10672 | 9702 | -0.013 | 0.2260 | No | ||
| 66 | GDF8 | 9411 | 9791 | -0.014 | 0.2216 | No | ||
| 67 | HNRPAB | 9104 | 9871 | -0.015 | 0.2177 | No | ||
| 68 | PRX | 3729 9628 | 10035 | -0.017 | 0.2093 | No | ||
| 69 | PRR3 | 22995 | 10062 | -0.018 | 0.2083 | No | ||
| 70 | PARP6 | 19419 2979 3156 | 10280 | -0.021 | 0.1970 | No | ||
| 71 | CSNK2A1 | 14797 | 10544 | -0.024 | 0.1834 | No | ||
| 72 | SPRY2 | 21725 | 11042 | -0.032 | 0.1573 | No | ||
| 73 | MEOX2 | 21285 | 11196 | -0.034 | 0.1498 | No | ||
| 74 | CUTL2 | 13110 4578 | 11545 | -0.040 | 0.1320 | No | ||
| 75 | SULF1 | 6285 | 11724 | -0.043 | 0.1233 | No | ||
| 76 | PCDHGA8 | 13548 | 11970 | -0.048 | 0.1112 | No | ||
| 77 | PCDHGB4 | 13534 | 11999 | -0.049 | 0.1109 | No | ||
| 78 | VAMP5 | 17118 | 12051 | -0.050 | 0.1093 | No | ||
| 79 | PCDHGB5 | 13535 | 12091 | -0.051 | 0.1084 | No | ||
| 80 | PCDHGA1 | 13541 | 12362 | -0.058 | 0.0952 | No | ||
| 81 | PCDHGB6 | 13536 | 12415 | -0.059 | 0.0937 | No | ||
| 82 | MLX | 958 1417 | 12417 | -0.059 | 0.0951 | No | ||
| 83 | PCDHGA2 | 13542 | 12462 | -0.060 | 0.0941 | No | ||
| 84 | HOXA1 | 1005 17152 | 12786 | -0.071 | 0.0783 | No | ||
| 85 | WDR1 | 16544 3592 | 12864 | -0.073 | 0.0759 | No | ||
| 86 | SEPT4 | 20713 | 13304 | -0.090 | 0.0543 | No | ||
| 87 | A2BP1 | 11212 1652 1689 | 13356 | -0.092 | 0.0537 | No | ||
| 88 | SYT7 | 3670 3698 23932 | 13513 | -0.100 | 0.0476 | No | ||
| 89 | GGA3 | 1206 6435 | 13630 | -0.106 | 0.0438 | No | ||
| 90 | ETV5 | 22630 | 14139 | -0.137 | 0.0196 | No | ||
| 91 | GRK5 | 9036 23814 | 14178 | -0.140 | 0.0208 | No | ||
| 92 | PCDHGA7 | 13547 | 14235 | -0.145 | 0.0212 | No | ||
| 93 | RND1 | 5868 | 14280 | -0.148 | 0.0223 | No | ||
| 94 | ING3 | 17514 1019 | 14300 | -0.150 | 0.0248 | No | ||
| 95 | TCF12 | 10044 3125 | 14586 | -0.178 | 0.0136 | No | ||
| 96 | ARID5B | 7728 | 14624 | -0.182 | 0.0158 | No | ||
| 97 | ZMYND11 | 12364 3269 3286 3223 | 14778 | -0.199 | 0.0122 | No | ||
| 98 | SALL2 | 21840 | 14785 | -0.200 | 0.0166 | No | ||
| 99 | ELAVL1 | 4888 | 14931 | -0.219 | 0.0139 | No | ||
| 100 | HMGN1 | 4856 | 15501 | -0.309 | -0.0096 | No | ||
| 101 | SLC30A4 | 14450 | 15831 | -0.385 | -0.0183 | No | ||
| 102 | PSME3 | 20657 | 16006 | -0.433 | -0.0175 | No | ||
| 103 | RHOT1 | 12227 | 16014 | -0.434 | -0.0077 | No | ||
| 104 | ST8SIA2 | 17797 | 16449 | -0.567 | -0.0178 | No | ||
| 105 | CSNK1G1 | 3057 5671 19397 | 16724 | -0.687 | -0.0165 | No | ||
| 106 | NR4A2 | 5203 | 16920 | -0.781 | -0.0087 | No | ||
| 107 | MAPK1 | 1642 11167 | 17667 | -1.160 | -0.0217 | No | ||
| 108 | DDT | 19732 | 17910 | -1.328 | -0.0036 | No | ||
| 109 | RALA | 21541 | 18299 | -1.773 | 0.0171 | No |