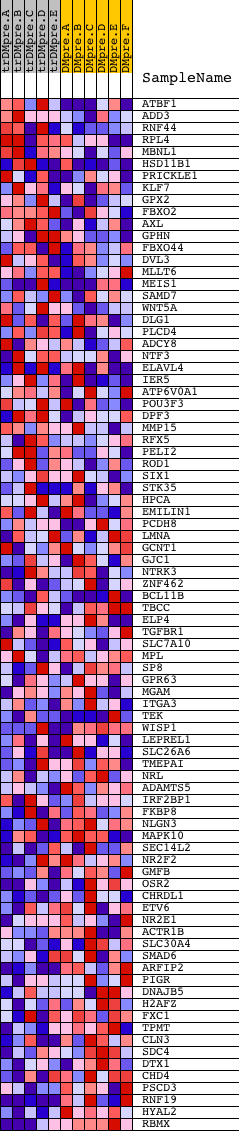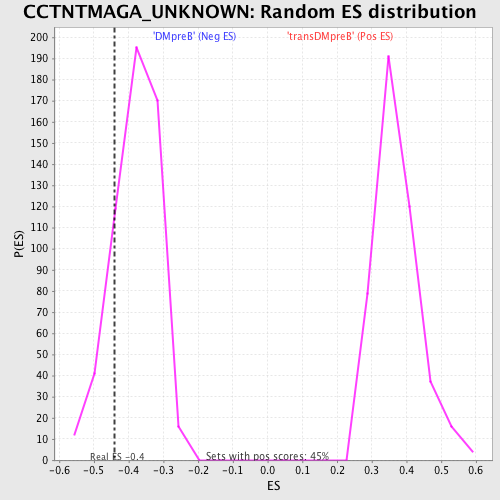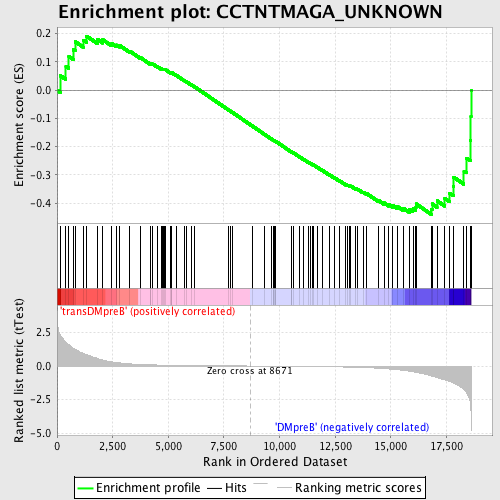
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | CCTNTMAGA_UNKNOWN |
| Enrichment Score (ES) | -0.43990856 |
| Normalized Enrichment Score (NES) | -1.1589277 |
| Nominal p-value | 0.16636528 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATBF1 | 916 8633 | 147 | 2.253 | 0.0513 | No | ||
| 2 | ADD3 | 23812 3694 | 397 | 1.777 | 0.0846 | No | ||
| 3 | RNF44 | 8364 | 529 | 1.588 | 0.1192 | No | ||
| 4 | RPL4 | 7499 19411 | 729 | 1.322 | 0.1433 | No | ||
| 5 | MBNL1 | 1921 15582 | 814 | 1.244 | 0.1714 | No | ||
| 6 | HSD11B1 | 9125 4019 | 1172 | 0.957 | 0.1773 | No | ||
| 7 | PRICKLE1 | 8379 | 1327 | 0.846 | 0.1912 | No | ||
| 8 | KLF7 | 8205 | 1819 | 0.553 | 0.1793 | No | ||
| 9 | GPX2 | 9037 | 2054 | 0.438 | 0.1782 | No | ||
| 10 | FBXO2 | 15993 | 2460 | 0.302 | 0.1643 | No | ||
| 11 | AXL | 17922 | 2667 | 0.256 | 0.1599 | No | ||
| 12 | GPHN | 6492 21232 | 2805 | 0.232 | 0.1586 | No | ||
| 13 | FBXO44 | 15675 | 3258 | 0.164 | 0.1385 | No | ||
| 14 | DVL3 | 8873 22821 | 3761 | 0.117 | 0.1145 | No | ||
| 15 | MLLT6 | 20678 | 4195 | 0.090 | 0.0935 | No | ||
| 16 | MEIS1 | 20524 | 4199 | 0.089 | 0.0957 | No | ||
| 17 | SAMD7 | 7998 13298 | 4295 | 0.084 | 0.0928 | No | ||
| 18 | WNT5A | 22066 5880 | 4522 | 0.075 | 0.0825 | No | ||
| 19 | DLG1 | 22793 1672 | 4679 | 0.069 | 0.0759 | No | ||
| 20 | PLCD4 | 9587 | 4756 | 0.066 | 0.0736 | No | ||
| 21 | ADCY8 | 22281 | 4797 | 0.065 | 0.0731 | No | ||
| 22 | NTF3 | 16991 | 4816 | 0.064 | 0.0739 | No | ||
| 23 | ELAVL4 | 15805 4889 9137 | 4859 | 0.063 | 0.0733 | No | ||
| 24 | IER5 | 13802 | 5111 | 0.055 | 0.0612 | No | ||
| 25 | ATP6V0A1 | 8640 4424 1197 | 5141 | 0.055 | 0.0611 | No | ||
| 26 | POU3F3 | 14261 | 5151 | 0.055 | 0.0620 | No | ||
| 27 | DPF3 | 7659 12853 | 5350 | 0.050 | 0.0526 | No | ||
| 28 | MMP15 | 18514 | 5709 | 0.042 | 0.0344 | No | ||
| 29 | RFX5 | 7018 | 5801 | 0.040 | 0.0305 | No | ||
| 30 | PELI2 | 8217 | 6042 | 0.035 | 0.0185 | No | ||
| 31 | ROD1 | 6047 2317 | 6181 | 0.033 | 0.0119 | No | ||
| 32 | SIX1 | 9821 | 7711 | 0.011 | -0.0703 | No | ||
| 33 | STK35 | 14851 | 7802 | 0.010 | -0.0749 | No | ||
| 34 | HPCA | 9118 | 7888 | 0.009 | -0.0793 | No | ||
| 35 | EMILIN1 | 16891 | 8786 | -0.001 | -0.1276 | No | ||
| 36 | PCDH8 | 21739 3017 | 9304 | -0.008 | -0.1553 | No | ||
| 37 | LMNA | 4999 15290 1778 9280 | 9619 | -0.012 | -0.1720 | No | ||
| 38 | GCNT1 | 23715 3749 3689 | 9720 | -0.013 | -0.1770 | No | ||
| 39 | GJC1 | 20256 | 9730 | -0.013 | -0.1771 | No | ||
| 40 | NTRK3 | 2069 3652 3705 9492 | 9764 | -0.014 | -0.1786 | No | ||
| 41 | ZNF462 | 6321 | 9821 | -0.014 | -0.1812 | No | ||
| 42 | BCL11B | 7190 | 9826 | -0.015 | -0.1810 | No | ||
| 43 | TBCC | 23215 | 10538 | -0.024 | -0.2188 | No | ||
| 44 | ELP4 | 8094 | 10556 | -0.025 | -0.2190 | No | ||
| 45 | TGFBR1 | 16213 10165 5745 | 10621 | -0.025 | -0.2218 | No | ||
| 46 | SLC7A10 | 18294 | 10916 | -0.030 | -0.2369 | No | ||
| 47 | MPL | 15780 | 11080 | -0.032 | -0.2449 | No | ||
| 48 | SP8 | 21122 | 11297 | -0.035 | -0.2556 | No | ||
| 49 | GPR63 | 16258 | 11367 | -0.037 | -0.2583 | No | ||
| 50 | MGAM | 17476 | 11489 | -0.039 | -0.2639 | No | ||
| 51 | ITGA3 | 20284 | 11505 | -0.039 | -0.2636 | No | ||
| 52 | TEK | 16175 | 11723 | -0.043 | -0.2742 | No | ||
| 53 | WISP1 | 22463 2289 | 11932 | -0.047 | -0.2842 | No | ||
| 54 | LEPREL1 | 5575 | 12245 | -0.055 | -0.2996 | No | ||
| 55 | SLC26A6 | 19304 | 12470 | -0.061 | -0.3101 | No | ||
| 56 | TMEPAI | 51 | 12700 | -0.068 | -0.3207 | No | ||
| 57 | NRL | 9484 | 12981 | -0.077 | -0.3337 | No | ||
| 58 | ADAMTS5 | 6207 | 13059 | -0.080 | -0.3358 | No | ||
| 59 | IRF2BP1 | 18368 | 13150 | -0.083 | -0.3384 | No | ||
| 60 | FKBP8 | 18587 | 13201 | -0.085 | -0.3389 | No | ||
| 61 | NLGN3 | 10878 | 13426 | -0.095 | -0.3485 | No | ||
| 62 | MAPK10 | 11169 | 13511 | -0.099 | -0.3504 | No | ||
| 63 | SEC14L2 | 20546 | 13751 | -0.112 | -0.3604 | No | ||
| 64 | NR2F2 | 8619 409 | 13896 | -0.121 | -0.3650 | No | ||
| 65 | GMFB | 21861 | 14439 | -0.164 | -0.3899 | No | ||
| 66 | OSR2 | 22492 | 14700 | -0.191 | -0.3989 | No | ||
| 67 | CHRDL1 | 8180 13504 | 14911 | -0.217 | -0.4045 | No | ||
| 68 | ETV6 | 17264 | 15086 | -0.238 | -0.4077 | No | ||
| 69 | NR2E1 | 19780 | 15278 | -0.269 | -0.4109 | No | ||
| 70 | ACTR1B | 30 | 15566 | -0.323 | -0.4179 | No | ||
| 71 | SLC30A4 | 14450 | 15831 | -0.385 | -0.4220 | Yes | ||
| 72 | SMAD6 | 19083 | 15996 | -0.430 | -0.4196 | Yes | ||
| 73 | ARFIP2 | 1226 17702 | 16125 | -0.465 | -0.4143 | Yes | ||
| 74 | PIGR | 9561 | 16135 | -0.467 | -0.4025 | Yes | ||
| 75 | DNAJB5 | 16236 | 16830 | -0.732 | -0.4207 | Yes | ||
| 76 | H2AFZ | 6957 | 16857 | -0.749 | -0.4024 | Yes | ||
| 77 | FXC1 | 8988 4043 | 17078 | -0.870 | -0.3914 | Yes | ||
| 78 | TPMT | 10213 | 17427 | -1.023 | -0.3833 | Yes | ||
| 79 | CLN3 | 17635 | 17648 | -1.147 | -0.3650 | Yes | ||
| 80 | SDC4 | 14349 | 17810 | -1.261 | -0.3405 | Yes | ||
| 81 | DTX1 | 16396 | 17823 | -1.270 | -0.3078 | Yes | ||
| 82 | CHD4 | 8418 17281 4225 | 18288 | -1.757 | -0.2867 | Yes | ||
| 83 | PSCD3 | 16633 | 18401 | -1.968 | -0.2410 | Yes | ||
| 84 | RNF19 | 22314 | 18574 | -2.785 | -0.1771 | Yes | ||
| 85 | HYAL2 | 4891 | 18601 | -3.278 | -0.0923 | Yes | ||
| 86 | RBMX | 5367 9708 | 18608 | -3.540 | 0.0004 | Yes |