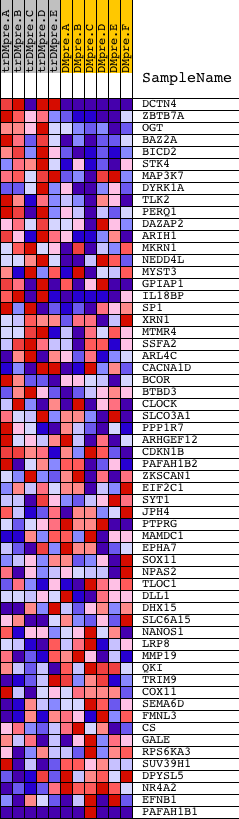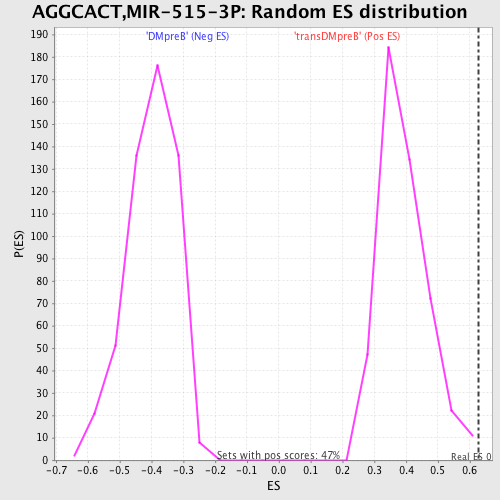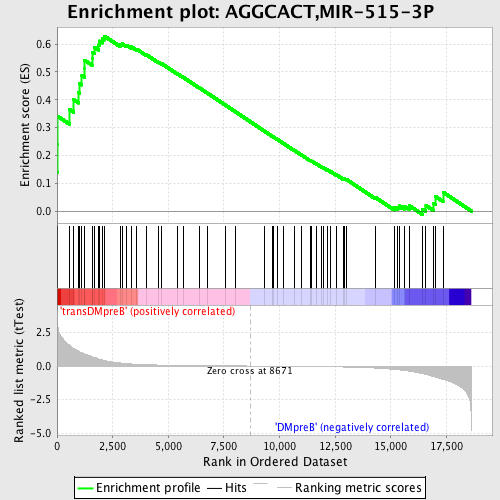
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | AGGCACT,MIR-515-3P |
| Enrichment Score (ES) | 0.6285115 |
| Normalized Enrichment Score (NES) | 1.6022388 |
| Nominal p-value | 0.0021276595 |
| FDR q-value | 0.22073358 |
| FWER p-Value | 0.369 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DCTN4 | 12560 7474 | 0 | 4.098 | 0.1403 | Yes | ||
| 2 | ZBTB7A | 5004 | 24 | 2.966 | 0.2406 | Yes | ||
| 3 | OGT | 4241 24274 | 32 | 2.930 | 0.3406 | Yes | ||
| 4 | BAZ2A | 4371 | 554 | 1.554 | 0.3657 | Yes | ||
| 5 | BICD2 | 8048 | 751 | 1.301 | 0.3997 | Yes | ||
| 6 | STK4 | 14743 | 965 | 1.084 | 0.4254 | Yes | ||
| 7 | MAP3K7 | 16255 | 1018 | 1.035 | 0.4580 | Yes | ||
| 8 | DYRK1A | 4649 | 1113 | 0.993 | 0.4869 | Yes | ||
| 9 | TLK2 | 20628 | 1238 | 0.905 | 0.5112 | Yes | ||
| 10 | PERQ1 | 7168 | 1241 | 0.904 | 0.5421 | Yes | ||
| 11 | DAZAP2 | 10758 | 1575 | 0.696 | 0.5479 | Yes | ||
| 12 | ARIH1 | 6210 | 1581 | 0.693 | 0.5714 | Yes | ||
| 13 | MKRN1 | 17178 | 1678 | 0.641 | 0.5882 | Yes | ||
| 14 | NEDD4L | 8192 | 1860 | 0.533 | 0.5967 | Yes | ||
| 15 | MYST3 | 18651 | 1913 | 0.500 | 0.6110 | Yes | ||
| 16 | GPIAP1 | 2819 7014 | 2049 | 0.439 | 0.6188 | Yes | ||
| 17 | IL18BP | 17730 | 2129 | 0.408 | 0.6285 | Yes | ||
| 18 | SP1 | 9852 | 2840 | 0.228 | 0.5981 | No | ||
| 19 | XRN1 | 10797 | 2921 | 0.212 | 0.6010 | No | ||
| 20 | MTMR4 | 20712 | 3130 | 0.182 | 0.5960 | No | ||
| 21 | SSFA2 | 7692 | 3330 | 0.155 | 0.5906 | No | ||
| 22 | ARL4C | 13888 | 3583 | 0.132 | 0.5816 | No | ||
| 23 | CACNA1D | 4464 8675 | 4027 | 0.100 | 0.5611 | No | ||
| 24 | BCOR | 12934 2561 | 4568 | 0.073 | 0.5345 | No | ||
| 25 | BTBD3 | 10457 2975 6007 6006 | 4703 | 0.068 | 0.5296 | No | ||
| 26 | CLOCK | 8749 16507 4530 | 5415 | 0.048 | 0.4929 | No | ||
| 27 | SLCO3A1 | 17796 4237 8430 | 5672 | 0.042 | 0.4806 | No | ||
| 28 | PPP1R7 | 14176 | 6416 | 0.029 | 0.4416 | No | ||
| 29 | ARHGEF12 | 19156 | 6766 | 0.024 | 0.4236 | No | ||
| 30 | CDKN1B | 4512 8730 | 7582 | 0.013 | 0.3801 | No | ||
| 31 | PAFAH1B2 | 5221 9525 | 8015 | 0.008 | 0.3571 | No | ||
| 32 | ZKSCAN1 | 7927 | 9314 | -0.008 | 0.2874 | No | ||
| 33 | EIF2C1 | 10672 | 9702 | -0.013 | 0.2670 | No | ||
| 34 | SYT1 | 5565 | 9706 | -0.013 | 0.2673 | No | ||
| 35 | JPH4 | 21826 | 9921 | -0.016 | 0.2563 | No | ||
| 36 | PTPRG | 22097 3151 | 10187 | -0.019 | 0.2426 | No | ||
| 37 | MAMDC1 | 21054 6798 | 10659 | -0.026 | 0.2182 | No | ||
| 38 | EPHA7 | 8909 4674 | 10969 | -0.030 | 0.2025 | No | ||
| 39 | SOX11 | 5477 | 11382 | -0.037 | 0.1816 | No | ||
| 40 | NPAS2 | 5186 | 11434 | -0.038 | 0.1801 | No | ||
| 41 | TLOC1 | 12787 | 11641 | -0.041 | 0.1705 | No | ||
| 42 | DLL1 | 23119 | 11894 | -0.047 | 0.1585 | No | ||
| 43 | DHX15 | 8842 | 11985 | -0.049 | 0.1553 | No | ||
| 44 | SLC6A15 | 8335 3321 | 12132 | -0.052 | 0.1492 | No | ||
| 45 | NANOS1 | 6858 | 12303 | -0.056 | 0.1420 | No | ||
| 46 | LRP8 | 16149 2425 | 12550 | -0.063 | 0.1309 | No | ||
| 47 | MMP19 | 7191 | 12874 | -0.073 | 0.1160 | No | ||
| 48 | QKI | 23128 5340 | 12921 | -0.075 | 0.1161 | No | ||
| 49 | TRIM9 | 2118 8076 13577 8234 | 13003 | -0.078 | 0.1144 | No | ||
| 50 | COX11 | 20705 | 14311 | -0.151 | 0.0491 | No | ||
| 51 | SEMA6D | 2800 14876 90 | 15148 | -0.248 | 0.0126 | No | ||
| 52 | FMNL3 | 22134 5866 | 15305 | -0.274 | 0.0136 | No | ||
| 53 | CS | 19839 | 15391 | -0.287 | 0.0188 | No | ||
| 54 | GALE | 13187 16032 | 15610 | -0.334 | 0.0185 | No | ||
| 55 | RPS6KA3 | 8490 | 15832 | -0.385 | 0.0197 | No | ||
| 56 | SUV39H1 | 24194 | 16422 | -0.559 | 0.0071 | No | ||
| 57 | DPYSL5 | 92 | 16579 | -0.624 | 0.0201 | No | ||
| 58 | NR4A2 | 5203 | 16920 | -0.781 | 0.0285 | No | ||
| 59 | EFNB1 | 24285 | 17011 | -0.834 | 0.0522 | No | ||
| 60 | PAFAH1B1 | 1340 5220 9524 | 17349 | -0.998 | 0.0683 | No |