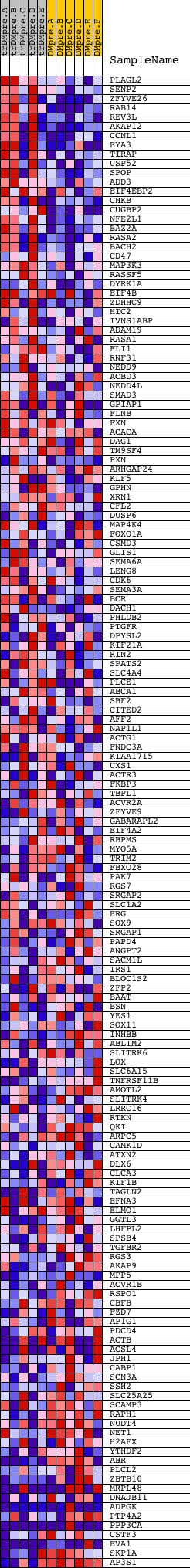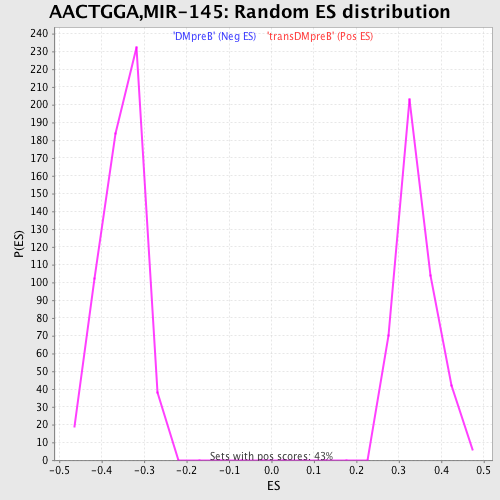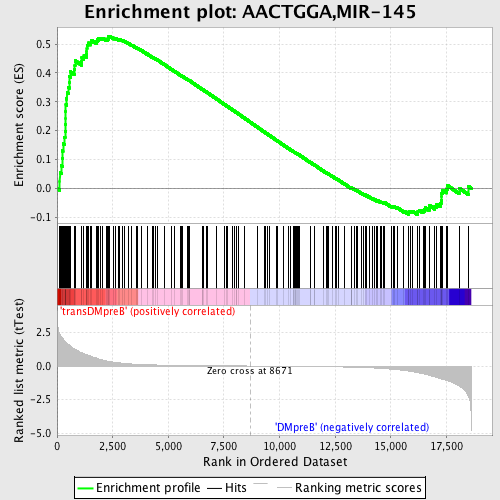
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | AACTGGA,MIR-145 |
| Enrichment Score (ES) | 0.5277613 |
| Normalized Enrichment Score (NES) | 1.5422192 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.17981704 |
| FWER p-Value | 0.646 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PLAGL2 | 7055 | 127 | 2.337 | 0.0235 | Yes | ||
| 2 | SENP2 | 7990 | 129 | 2.334 | 0.0539 | Yes | ||
| 3 | ZFYVE26 | 9999 | 195 | 2.169 | 0.0786 | Yes | ||
| 4 | RAB14 | 12685 | 232 | 2.087 | 0.1038 | Yes | ||
| 5 | REV3L | 20050 | 242 | 2.072 | 0.1303 | Yes | ||
| 6 | AKAP12 | 17600 | 266 | 2.027 | 0.1554 | Yes | ||
| 7 | CCNL1 | 15325 | 323 | 1.872 | 0.1767 | Yes | ||
| 8 | EYA3 | 8936 2352 | 358 | 1.821 | 0.1986 | Yes | ||
| 9 | TIRAP | 3075 19187 | 378 | 1.797 | 0.2210 | Yes | ||
| 10 | USP52 | 3397 19841 | 387 | 1.783 | 0.2437 | Yes | ||
| 11 | SPOP | 5498 | 390 | 1.780 | 0.2668 | Yes | ||
| 12 | ADD3 | 23812 3694 | 397 | 1.777 | 0.2896 | Yes | ||
| 13 | EIF4EBP2 | 4662 | 430 | 1.718 | 0.3102 | Yes | ||
| 14 | CHKB | 4518 | 444 | 1.707 | 0.3318 | Yes | ||
| 15 | CUGBP2 | 4687 8923 | 502 | 1.629 | 0.3499 | Yes | ||
| 16 | NFE2L1 | 9457 | 545 | 1.563 | 0.3679 | Yes | ||
| 17 | BAZ2A | 4371 | 554 | 1.554 | 0.3877 | Yes | ||
| 18 | RASA2 | 4329 | 608 | 1.476 | 0.4041 | Yes | ||
| 19 | BACH2 | 8648 | 769 | 1.283 | 0.4121 | Yes | ||
| 20 | CD47 | 4933 | 801 | 1.260 | 0.4268 | Yes | ||
| 21 | MAP3K3 | 20626 | 808 | 1.254 | 0.4428 | Yes | ||
| 22 | RASSF5 | 13837 | 1100 | 0.998 | 0.4400 | Yes | ||
| 23 | DYRK1A | 4649 | 1113 | 0.993 | 0.4523 | Yes | ||
| 24 | EIF4B | 13279 7979 | 1207 | 0.924 | 0.4593 | Yes | ||
| 25 | ZDHHC9 | 24163 | 1320 | 0.849 | 0.4643 | Yes | ||
| 26 | HIC2 | 7185 | 1337 | 0.842 | 0.4744 | Yes | ||
| 27 | IVNS1ABP | 4384 4050 | 1342 | 0.840 | 0.4851 | Yes | ||
| 28 | ADAM19 | 8548 | 1345 | 0.838 | 0.4959 | Yes | ||
| 29 | RASA1 | 10174 | 1390 | 0.808 | 0.5040 | Yes | ||
| 30 | FLI1 | 4729 | 1522 | 0.726 | 0.5064 | Yes | ||
| 31 | RNF31 | 11206 | 1550 | 0.706 | 0.5141 | Yes | ||
| 32 | NEDD9 | 3206 21483 | 1777 | 0.580 | 0.5094 | Yes | ||
| 33 | ACBD3 | 14025 | 1812 | 0.556 | 0.5148 | Yes | ||
| 34 | NEDD4L | 8192 | 1860 | 0.533 | 0.5192 | Yes | ||
| 35 | SMAD3 | 19084 | 1948 | 0.484 | 0.5208 | Yes | ||
| 36 | GPIAP1 | 2819 7014 | 2049 | 0.439 | 0.5211 | Yes | ||
| 37 | FLNB | 13742 | 2215 | 0.378 | 0.5171 | Yes | ||
| 38 | FXN | 23709 | 2285 | 0.358 | 0.5180 | Yes | ||
| 39 | ACACA | 309 4209 | 2298 | 0.356 | 0.5220 | Yes | ||
| 40 | DAG1 | 18996 8837 | 2306 | 0.353 | 0.5262 | Yes | ||
| 41 | TM9SF4 | 14786 2830 8279 | 2358 | 0.334 | 0.5278 | Yes | ||
| 42 | PXN | 5339 3573 | 2545 | 0.281 | 0.5213 | No | ||
| 43 | ARHGAP24 | 16778 3655 | 2616 | 0.266 | 0.5210 | No | ||
| 44 | KLF5 | 4456 | 2761 | 0.240 | 0.5163 | No | ||
| 45 | GPHN | 6492 21232 | 2805 | 0.232 | 0.5170 | No | ||
| 46 | XRN1 | 10797 | 2921 | 0.212 | 0.5135 | No | ||
| 47 | CFL2 | 2155 21069 | 3023 | 0.197 | 0.5106 | No | ||
| 48 | DUSP6 | 19891 3399 | 3198 | 0.172 | 0.5035 | No | ||
| 49 | MAP4K4 | 11241 | 3363 | 0.151 | 0.4965 | No | ||
| 50 | FOXO1A | 7124 | 3563 | 0.134 | 0.4875 | No | ||
| 51 | CSMD3 | 10735 7938 | 3626 | 0.129 | 0.4858 | No | ||
| 52 | GLIS1 | 16150 | 3774 | 0.116 | 0.4794 | No | ||
| 53 | SEMA6A | 23439 9802 | 4042 | 0.100 | 0.4662 | No | ||
| 54 | LENG8 | 6114 | 4284 | 0.085 | 0.4542 | No | ||
| 55 | CDK6 | 16600 | 4318 | 0.083 | 0.4535 | No | ||
| 56 | SEMA3A | 16919 | 4428 | 0.079 | 0.4487 | No | ||
| 57 | BCR | 8478 3384 19990 | 4523 | 0.075 | 0.4445 | No | ||
| 58 | DACH1 | 4595 3371 | 4807 | 0.065 | 0.4300 | No | ||
| 59 | PHLDB2 | 22581 | 5162 | 0.054 | 0.4116 | No | ||
| 60 | PTGFR | 15138 | 5284 | 0.051 | 0.4057 | No | ||
| 61 | DPYSL2 | 3122 4561 8787 | 5551 | 0.045 | 0.3918 | No | ||
| 62 | KIF21A | 4952 | 5600 | 0.044 | 0.3898 | No | ||
| 63 | RIN2 | 14817 | 5626 | 0.043 | 0.3890 | No | ||
| 64 | SPATS2 | 13044 | 5873 | 0.038 | 0.3762 | No | ||
| 65 | SLC4A4 | 16807 12035 7036 | 5912 | 0.037 | 0.3746 | No | ||
| 66 | PLCE1 | 13156 | 5934 | 0.037 | 0.3740 | No | ||
| 67 | ABCA1 | 15881 | 5965 | 0.036 | 0.3728 | No | ||
| 68 | SBF2 | 11485 | 6525 | 0.027 | 0.3429 | No | ||
| 69 | CITED2 | 5118 14477 | 6554 | 0.027 | 0.3417 | No | ||
| 70 | AFF2 | 8977 | 6578 | 0.027 | 0.3408 | No | ||
| 71 | NAP1L1 | 19882 3427 330 9485 5193 | 6703 | 0.024 | 0.3344 | No | ||
| 72 | ACTG1 | 8535 | 6767 | 0.024 | 0.3313 | No | ||
| 73 | FNDC3A | 6672 21756 | 7142 | 0.018 | 0.3113 | No | ||
| 74 | KIAA1715 | 7629 | 7526 | 0.013 | 0.2907 | No | ||
| 75 | UXS1 | 13965 | 7602 | 0.012 | 0.2868 | No | ||
| 76 | ACTR3 | 13166 | 7669 | 0.012 | 0.2834 | No | ||
| 77 | FKBP3 | 21056 | 7864 | 0.009 | 0.2730 | No | ||
| 78 | TBPL1 | 19805 | 7956 | 0.008 | 0.2681 | No | ||
| 79 | ACVR2A | 8542 | 8040 | 0.007 | 0.2637 | No | ||
| 80 | ZFYVE9 | 10501 2449 | 8057 | 0.007 | 0.2630 | No | ||
| 81 | GABARAPL2 | 8211 13552 | 8166 | 0.006 | 0.2572 | No | ||
| 82 | EIF4A2 | 4660 1679 1645 | 8430 | 0.003 | 0.2430 | No | ||
| 83 | RBPMS | 5369 | 9021 | -0.004 | 0.2111 | No | ||
| 84 | MYO5A | 19378 2978 | 9315 | -0.008 | 0.1953 | No | ||
| 85 | TRIM2 | 13485 8157 | 9351 | -0.008 | 0.1935 | No | ||
| 86 | FBXO28 | 7508 12612 | 9460 | -0.010 | 0.1878 | No | ||
| 87 | PAK7 | 14417 | 9538 | -0.011 | 0.1837 | No | ||
| 88 | RGS7 | 13743 | 9852 | -0.015 | 0.1670 | No | ||
| 89 | SRGAP2 | 13836 17045 6433 17046 | 9902 | -0.016 | 0.1645 | No | ||
| 90 | SLC1A2 | 5453 | 10174 | -0.019 | 0.1501 | No | ||
| 91 | ERG | 1686 8915 | 10402 | -0.022 | 0.1381 | No | ||
| 92 | SOX9 | 20611 | 10472 | -0.023 | 0.1346 | No | ||
| 93 | SRGAP1 | 19615 | 10602 | -0.025 | 0.1280 | No | ||
| 94 | PAPD4 | 8299 8300 4146 | 10625 | -0.026 | 0.1271 | No | ||
| 95 | ANGPT2 | 14885 3783 18923 | 10662 | -0.026 | 0.1255 | No | ||
| 96 | SACM1L | 19253 | 10708 | -0.027 | 0.1234 | No | ||
| 97 | IRS1 | 4925 | 10717 | -0.027 | 0.1233 | No | ||
| 98 | BLOC1S2 | 23664 | 10751 | -0.027 | 0.1219 | No | ||
| 99 | ZFP2 | 10385 1366 1474 | 10806 | -0.028 | 0.1193 | No | ||
| 100 | BAAT | 15885 | 10847 | -0.029 | 0.1175 | No | ||
| 101 | BSN | 18997 | 10883 | -0.029 | 0.1160 | No | ||
| 102 | YES1 | 5930 | 11369 | -0.037 | 0.0902 | No | ||
| 103 | SOX11 | 5477 | 11382 | -0.037 | 0.0900 | No | ||
| 104 | INHBB | 13858 | 11553 | -0.040 | 0.0814 | No | ||
| 105 | ABLIM2 | 16866 3605 | 11586 | -0.041 | 0.0801 | No | ||
| 106 | SLITRK6 | 21724 | 11958 | -0.048 | 0.0607 | No | ||
| 107 | LOX | 23438 | 12086 | -0.051 | 0.0545 | No | ||
| 108 | SLC6A15 | 8335 3321 | 12132 | -0.052 | 0.0527 | No | ||
| 109 | TNFRSF11B | 22296 | 12199 | -0.054 | 0.0498 | No | ||
| 110 | AMOTL2 | 19339 | 12382 | -0.058 | 0.0407 | No | ||
| 111 | SLITRK4 | 24144 | 12493 | -0.061 | 0.0356 | No | ||
| 112 | LRRC16 | 21515 21514 | 12542 | -0.063 | 0.0338 | No | ||
| 113 | RTKN | 17394 1156 | 12647 | -0.066 | 0.0290 | No | ||
| 114 | QKI | 23128 5340 | 12921 | -0.075 | 0.0152 | No | ||
| 115 | ARPC5 | 12580 7487 | 13211 | -0.086 | 0.0006 | No | ||
| 116 | CAMK1D | 5977 2948 | 13212 | -0.086 | 0.0018 | No | ||
| 117 | ATXN2 | 9780 3506 | 13229 | -0.086 | 0.0020 | No | ||
| 118 | DLX6 | 17527 | 13362 | -0.092 | -0.0039 | No | ||
| 119 | CLCA3 | 6224 | 13452 | -0.096 | -0.0075 | No | ||
| 120 | KIF1B | 15671 2476 4950 15673 | 13486 | -0.098 | -0.0080 | No | ||
| 121 | TAGLN2 | 14043 | 13667 | -0.108 | -0.0164 | No | ||
| 122 | EFNA3 | 15278 | 13781 | -0.114 | -0.0210 | No | ||
| 123 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 13839 | -0.118 | -0.0226 | No | ||
| 124 | GGTL3 | 14381 | 13924 | -0.122 | -0.0255 | No | ||
| 125 | LHFPL2 | 21584 | 14035 | -0.130 | -0.0298 | No | ||
| 126 | SPSB4 | 19035 | 14180 | -0.140 | -0.0358 | No | ||
| 127 | TGFBR2 | 2994 3001 10167 | 14270 | -0.147 | -0.0387 | No | ||
| 128 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 14377 | -0.157 | -0.0424 | No | ||
| 129 | AKAP9 | 16604 16603 4150 8303 | 14383 | -0.158 | -0.0406 | No | ||
| 130 | MPP5 | 21230 7078 | 14546 | -0.174 | -0.0471 | No | ||
| 131 | ACVR1B | 4335 22353 | 14601 | -0.180 | -0.0477 | No | ||
| 132 | RSPO1 | 16087 | 14684 | -0.190 | -0.0497 | No | ||
| 133 | CBFB | 3769 18499 | 14715 | -0.192 | -0.0488 | No | ||
| 134 | FZD7 | 14242 | 15014 | -0.230 | -0.0620 | No | ||
| 135 | AP1G1 | 4392 | 15128 | -0.245 | -0.0649 | No | ||
| 136 | PDCD4 | 5232 23816 | 15152 | -0.248 | -0.0629 | No | ||
| 137 | ACTB | 8534 337 337 338 | 15280 | -0.269 | -0.0663 | No | ||
| 138 | ACSL4 | 24043 11936 24042 2614 6940 | 15587 | -0.328 | -0.0786 | No | ||
| 139 | JPH1 | 13994 | 15799 | -0.377 | -0.0851 | No | ||
| 140 | CABP1 | 16412 | 15815 | -0.380 | -0.0810 | No | ||
| 141 | SCN3A | 14573 | 15884 | -0.398 | -0.0795 | No | ||
| 142 | SSH2 | 6202 | 15972 | -0.423 | -0.0787 | No | ||
| 143 | SLC25A25 | 10431 | 16194 | -0.485 | -0.0844 | No | ||
| 144 | SCAMP3 | 15543 | 16216 | -0.490 | -0.0791 | No | ||
| 145 | RAPH1 | 19110 | 16268 | -0.504 | -0.0753 | No | ||
| 146 | NUDT4 | 19639 | 16451 | -0.567 | -0.0778 | No | ||
| 147 | NET1 | 7100 | 16515 | -0.593 | -0.0735 | No | ||
| 148 | H2AFX | 19477 | 16548 | -0.610 | -0.0673 | No | ||
| 149 | YTHDF2 | 5623 5624 | 16720 | -0.684 | -0.0677 | No | ||
| 150 | ABR | 8468 20346 1476 1429 | 16746 | -0.697 | -0.0600 | No | ||
| 151 | PLCL2 | 5902 | 16977 | -0.815 | -0.0618 | No | ||
| 152 | ZBTB10 | 10468 | 17066 | -0.863 | -0.0553 | No | ||
| 153 | MRPL48 | 3328 17739 2035 2242 3883 | 17236 | -0.942 | -0.0522 | No | ||
| 154 | DNAJB11 | 22813 | 17262 | -0.957 | -0.0411 | No | ||
| 155 | ADPGK | 13003 7788 19422 | 17284 | -0.968 | -0.0297 | No | ||
| 156 | PTP4A2 | 9659 2463 2518 5324 | 17288 | -0.972 | -0.0172 | No | ||
| 157 | PPP3CA | 1863 5284 | 17331 | -0.993 | -0.0065 | No | ||
| 158 | CSTF3 | 10449 6002 | 17511 | -1.069 | -0.0023 | No | ||
| 159 | EVA1 | 19472 | 17538 | -1.081 | 0.0103 | No | ||
| 160 | SKP1A | 20889 | 18072 | -1.475 | 0.0007 | No | ||
| 161 | AP3S1 | 8604 | 18480 | -2.210 | 0.0074 | No |