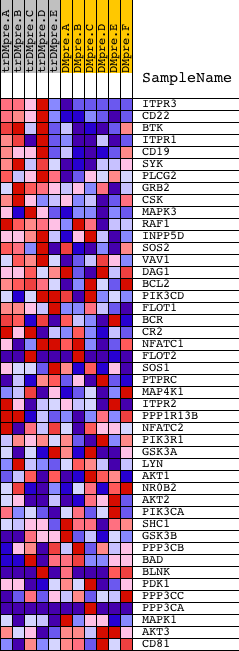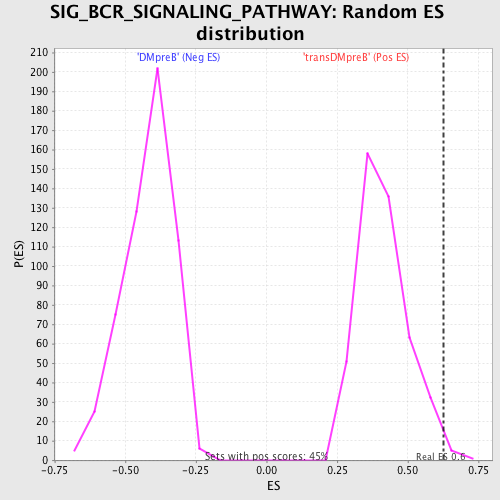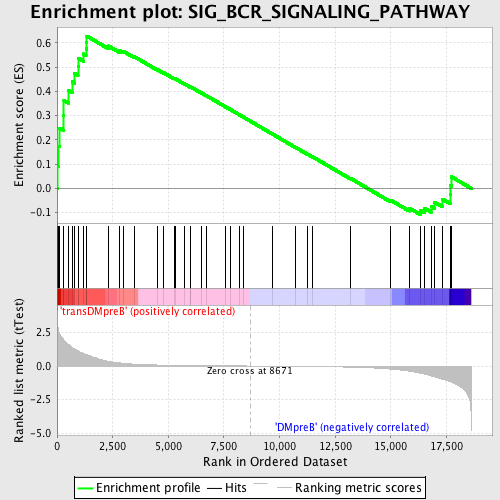
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | SIG_BCR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | 0.62801903 |
| Normalized Enrichment Score (NES) | 1.5242095 |
| Nominal p-value | 0.00896861 |
| FDR q-value | 0.29397157 |
| FWER p-Value | 0.981 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ITPR3 | 9195 | 31 | 2.936 | 0.0926 | Yes | ||
| 2 | CD22 | 17881 | 70 | 2.621 | 0.1748 | Yes | ||
| 3 | BTK | 24061 | 118 | 2.378 | 0.2487 | Yes | ||
| 4 | ITPR1 | 17341 | 285 | 1.959 | 0.3027 | Yes | ||
| 5 | CD19 | 17640 | 306 | 1.909 | 0.3629 | Yes | ||
| 6 | SYK | 21636 | 513 | 1.619 | 0.4039 | Yes | ||
| 7 | PLCG2 | 18453 | 681 | 1.392 | 0.4396 | Yes | ||
| 8 | GRB2 | 20149 | 792 | 1.266 | 0.4744 | Yes | ||
| 9 | CSK | 8805 | 940 | 1.117 | 0.5023 | Yes | ||
| 10 | MAPK3 | 6458 11170 | 961 | 1.090 | 0.5362 | Yes | ||
| 11 | RAF1 | 17035 | 1186 | 0.942 | 0.5545 | Yes | ||
| 12 | INPP5D | 14198 | 1301 | 0.857 | 0.5759 | Yes | ||
| 13 | SOS2 | 21049 | 1331 | 0.845 | 0.6014 | Yes | ||
| 14 | VAV1 | 23173 | 1340 | 0.841 | 0.6280 | Yes | ||
| 15 | DAG1 | 18996 8837 | 2306 | 0.353 | 0.5874 | No | ||
| 16 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 2820 | 0.230 | 0.5672 | No | ||
| 17 | PIK3CD | 9563 | 2973 | 0.204 | 0.5655 | No | ||
| 18 | FLOT1 | 23249 1613 | 3461 | 0.141 | 0.5439 | No | ||
| 19 | BCR | 8478 3384 19990 | 4523 | 0.075 | 0.4891 | No | ||
| 20 | CR2 | 3942 13707 | 4795 | 0.065 | 0.4766 | No | ||
| 21 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 5266 | 0.052 | 0.4530 | No | ||
| 22 | FLOT2 | 1247 8975 1430 | 5306 | 0.051 | 0.4525 | No | ||
| 23 | SOS1 | 5476 | 5727 | 0.041 | 0.4312 | No | ||
| 24 | PTPRC | 5327 9662 | 5973 | 0.036 | 0.4192 | No | ||
| 25 | MAP4K1 | 18313 | 6010 | 0.036 | 0.4184 | No | ||
| 26 | ITPR2 | 9194 | 6505 | 0.028 | 0.3927 | No | ||
| 27 | PPP1R13B | 10211 20980 | 6706 | 0.024 | 0.3827 | No | ||
| 28 | NFATC2 | 5168 2866 | 7581 | 0.013 | 0.3360 | No | ||
| 29 | PIK3R1 | 3170 | 7780 | 0.010 | 0.3257 | No | ||
| 30 | GSK3A | 405 | 8181 | 0.006 | 0.3043 | No | ||
| 31 | LYN | 16281 | 8372 | 0.004 | 0.2942 | No | ||
| 32 | AKT1 | 8568 | 9662 | -0.013 | 0.2252 | No | ||
| 33 | NR0B2 | 16050 | 10732 | -0.027 | 0.1685 | No | ||
| 34 | AKT2 | 4365 4366 | 11247 | -0.035 | 0.1419 | No | ||
| 35 | PIK3CA | 9562 | 11493 | -0.039 | 0.1300 | No | ||
| 36 | SHC1 | 9813 9812 5430 | 13187 | -0.085 | 0.0416 | No | ||
| 37 | GSK3B | 22761 | 14988 | -0.227 | -0.0481 | No | ||
| 38 | PPP3CB | 5285 | 15826 | -0.383 | -0.0808 | No | ||
| 39 | BAD | 24000 | 16322 | -0.526 | -0.0906 | No | ||
| 40 | BLNK | 23681 3691 | 16514 | -0.592 | -0.0818 | No | ||
| 41 | PDK1 | 14987 | 16809 | -0.726 | -0.0744 | No | ||
| 42 | PPP3CC | 21763 | 16969 | -0.809 | -0.0569 | No | ||
| 43 | PPP3CA | 1863 5284 | 17331 | -0.993 | -0.0445 | No | ||
| 44 | MAPK1 | 1642 11167 | 17667 | -1.160 | -0.0252 | No | ||
| 45 | AKT3 | 13739 982 | 17683 | -1.172 | 0.0116 | No | ||
| 46 | CD81 | 8719 | 17724 | -1.201 | 0.0480 | No |