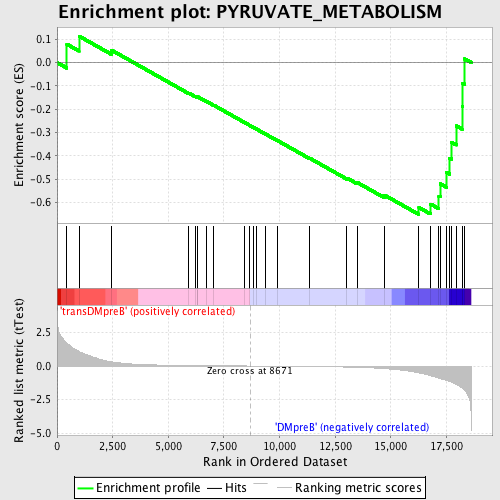
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | PYRUVATE_METABOLISM |
| Enrichment Score (ES) | -0.6520319 |
| Normalized Enrichment Score (NES) | -1.4248908 |
| Nominal p-value | 0.03954802 |
| FDR q-value | 0.3375354 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PKM2 | 3642 9573 | 439 | 1.709 | 0.0790 | No | ||
| 2 | ALDH2 | 16384 | 985 | 1.065 | 0.1137 | No | ||
| 3 | MDH2 | 9399 | 2446 | 0.306 | 0.0535 | No | ||
| 4 | PDHA2 | 15153 | 5919 | 0.037 | -0.1311 | No | ||
| 5 | ALDH3A1 | 20854 | 6209 | 0.032 | -0.1447 | No | ||
| 6 | ALDH1A1 | 8569 | 6305 | 0.031 | -0.1479 | No | ||
| 7 | ALDH1A2 | 19386 | 6309 | 0.031 | -0.1462 | No | ||
| 8 | LDHD | 11980 6993 | 6728 | 0.024 | -0.1673 | No | ||
| 9 | PKLR | 1850 15545 | 7037 | 0.020 | -0.1827 | No | ||
| 10 | PCK1 | 5228 | 8439 | 0.003 | -0.2579 | No | ||
| 11 | LDHC | 18235 2235 | 8640 | 0.000 | -0.2686 | No | ||
| 12 | ACYP2 | 20508 | 8836 | -0.002 | -0.2790 | No | ||
| 13 | ALDH9A1 | 14064 | 8980 | -0.004 | -0.2865 | No | ||
| 14 | HAGHL | 1528 7593 | 9364 | -0.009 | -0.3066 | No | ||
| 15 | ALDH1A3 | 17802 | 9918 | -0.016 | -0.3354 | No | ||
| 16 | PDHB | 12670 7548 | 11347 | -0.036 | -0.4100 | No | ||
| 17 | ACYP1 | 21019 | 13025 | -0.079 | -0.4955 | No | ||
| 18 | ACAT1 | 19114 10019 | 13487 | -0.098 | -0.5144 | No | ||
| 19 | MDH1 | 1245 9400 | 14722 | -0.193 | -0.5692 | No | ||
| 20 | ADH5 | 8554 15404 | 16262 | -0.501 | -0.6219 | Yes | ||
| 21 | DLAT | 19123 | 16784 | -0.713 | -0.6071 | Yes | ||
| 22 | DLD | 2097 21090 | 17160 | -0.902 | -0.5731 | Yes | ||
| 23 | PDHA1 | 24020 | 17220 | -0.933 | -0.5202 | Yes | ||
| 24 | ACAT2 | 8484 1537 | 17508 | -1.068 | -0.4715 | Yes | ||
| 25 | LDHA | 9269 | 17646 | -1.146 | -0.4101 | Yes | ||
| 26 | GLO1 | 23039 | 17730 | -1.205 | -0.3422 | Yes | ||
| 27 | ME2 | 4202 | 17947 | -1.366 | -0.2717 | Yes | ||
| 28 | HAGH | 9020 | 18207 | -1.630 | -0.1877 | Yes | ||
| 29 | ALDH1B1 | 16219 | 18233 | -1.672 | -0.0887 | Yes | ||
| 30 | GRHPR | 16224 | 18322 | -1.818 | 0.0158 | Yes |

