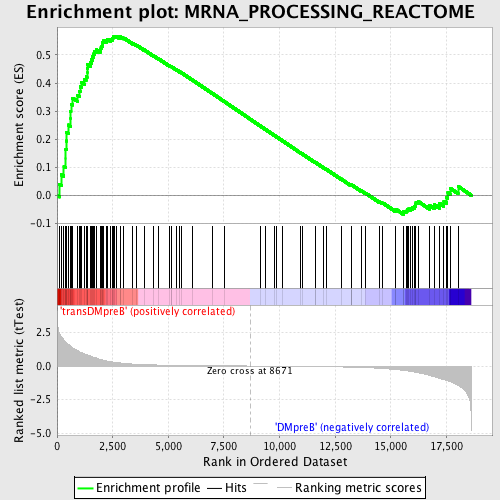
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | MRNA_PROCESSING_REACTOME |
| Enrichment Score (ES) | 0.5670902 |
| Normalized Enrichment Score (NES) | 1.5896351 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.29297462 |
| FWER p-Value | 0.814 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SFRS14 | 10622 | 111 | 2.412 | 0.0382 | Yes | ||
| 2 | CLK4 | 20895 1292 | 197 | 2.165 | 0.0733 | Yes | ||
| 3 | NXF1 | 11986 | 308 | 1.907 | 0.1023 | Yes | ||
| 4 | SPOP | 5498 | 390 | 1.780 | 0.1306 | Yes | ||
| 5 | SF3B2 | 23974 | 391 | 1.779 | 0.1632 | Yes | ||
| 6 | PABPN1 | 12018 | 416 | 1.736 | 0.1937 | Yes | ||
| 7 | SFRS5 | 9808 2062 | 426 | 1.724 | 0.2248 | Yes | ||
| 8 | CUGBP2 | 4687 8923 | 502 | 1.629 | 0.2506 | Yes | ||
| 9 | FUS | 6135 | 599 | 1.491 | 0.2728 | Yes | ||
| 10 | SRRM1 | 6958 | 605 | 1.481 | 0.2997 | Yes | ||
| 11 | CPSF1 | 22241 | 625 | 1.454 | 0.3253 | Yes | ||
| 12 | CD2BP2 | 17621 | 696 | 1.369 | 0.3466 | Yes | ||
| 13 | PRPF8 | 20780 1371 | 925 | 1.138 | 0.3551 | Yes | ||
| 14 | DHX38 | 48 | 1006 | 1.047 | 0.3700 | Yes | ||
| 15 | DHX9 | 8845 4608 3948 | 1037 | 1.022 | 0.3871 | Yes | ||
| 16 | PTBP1 | 5303 | 1089 | 1.000 | 0.4027 | Yes | ||
| 17 | SUPT5H | 9939 | 1232 | 0.907 | 0.4117 | Yes | ||
| 18 | SF3A1 | 7450 | 1303 | 0.856 | 0.4236 | Yes | ||
| 19 | SNRPF | 7645 | 1348 | 0.835 | 0.4365 | Yes | ||
| 20 | DHX8 | 20649 | 1353 | 0.831 | 0.4516 | Yes | ||
| 21 | SRPK1 | 23049 | 1373 | 0.817 | 0.4655 | Yes | ||
| 22 | RBM17 | 13371 | 1494 | 0.746 | 0.4727 | Yes | ||
| 23 | HNRPU | 6959 | 1547 | 0.710 | 0.4829 | Yes | ||
| 24 | SNRPD3 | 12514 | 1586 | 0.689 | 0.4935 | Yes | ||
| 25 | DDX1 | 21104 | 1627 | 0.665 | 0.5035 | Yes | ||
| 26 | DHX16 | 23247 | 1688 | 0.636 | 0.5119 | Yes | ||
| 27 | NONO | 11993 | 1759 | 0.593 | 0.5190 | Yes | ||
| 28 | SNRP70 | 1186 | 1933 | 0.488 | 0.5186 | Yes | ||
| 29 | SFRS2 | 9807 20136 | 1971 | 0.471 | 0.5253 | Yes | ||
| 30 | PRPF3 | 12899 12898 1877 7703 | 2010 | 0.454 | 0.5315 | Yes | ||
| 31 | SF4 | 18593 | 2038 | 0.442 | 0.5382 | Yes | ||
| 32 | GIPC1 | 3841 3882 18552 | 2048 | 0.440 | 0.5458 | Yes | ||
| 33 | DICER1 | 20989 | 2098 | 0.420 | 0.5508 | Yes | ||
| 34 | SF3B4 | 22269 | 2210 | 0.381 | 0.5518 | Yes | ||
| 35 | DDIT3 | 19857 | 2251 | 0.369 | 0.5564 | Yes | ||
| 36 | SNRPB | 9842 5469 2736 | 2403 | 0.322 | 0.5542 | Yes | ||
| 37 | CLK2 | 1883 15544 | 2477 | 0.298 | 0.5557 | Yes | ||
| 38 | HNRPR | 13196 7909 2500 | 2499 | 0.294 | 0.5599 | Yes | ||
| 39 | PRPF4 | 7655 12845 | 2515 | 0.288 | 0.5644 | Yes | ||
| 40 | SFRS9 | 16731 | 2565 | 0.277 | 0.5669 | Yes | ||
| 41 | HNRPA3 | 6023 2712 10473 2683 | 2650 | 0.260 | 0.5671 | Yes | ||
| 42 | CUGBP1 | 2805 8819 4576 2924 | 2834 | 0.228 | 0.5614 | No | ||
| 43 | PPM1G | 16571 | 2847 | 0.226 | 0.5649 | No | ||
| 44 | U2AF1 | 8431 | 2998 | 0.200 | 0.5605 | No | ||
| 45 | HNRPH2 | 12085 7082 | 3403 | 0.147 | 0.5413 | No | ||
| 46 | SFRS6 | 14751 | 3569 | 0.133 | 0.5349 | No | ||
| 47 | U2AF2 | 5813 5812 5814 | 3912 | 0.107 | 0.5184 | No | ||
| 48 | PHF5A | 22194 | 4338 | 0.082 | 0.4969 | No | ||
| 49 | SNRPN | 1715 9844 5471 | 4567 | 0.073 | 0.4860 | No | ||
| 50 | SFRS8 | 10543 6089 | 5046 | 0.057 | 0.4612 | No | ||
| 51 | RNPS1 | 9730 23361 | 5119 | 0.055 | 0.4583 | No | ||
| 52 | RBM5 | 13507 3016 | 5134 | 0.055 | 0.4586 | No | ||
| 53 | SF3A3 | 16091 | 5382 | 0.049 | 0.4461 | No | ||
| 54 | PAPOLA | 5261 9585 | 5521 | 0.045 | 0.4395 | No | ||
| 55 | NUDT21 | 12665 | 5568 | 0.044 | 0.4379 | No | ||
| 56 | SNRPB2 | 5470 | 6101 | 0.034 | 0.4098 | No | ||
| 57 | SFRS16 | 17943 3840 | 6991 | 0.020 | 0.3621 | No | ||
| 58 | RNGTT | 10769 2354 6272 | 7529 | 0.013 | 0.3334 | No | ||
| 59 | SFRS10 | 9818 1722 5441 | 9130 | -0.005 | 0.2471 | No | ||
| 60 | CPSF3 | 7038 | 9369 | -0.009 | 0.2344 | No | ||
| 61 | CSTF2T | 13502 | 9759 | -0.014 | 0.2136 | No | ||
| 62 | HNRPAB | 9104 | 9871 | -0.015 | 0.2079 | No | ||
| 63 | COL2A1 | 22145 4542 | 10131 | -0.019 | 0.1943 | No | ||
| 64 | HNRPA2B1 | 1187 7001 7000 1040 | 10954 | -0.030 | 0.1504 | No | ||
| 65 | HNRPD | 8645 | 11009 | -0.031 | 0.1481 | No | ||
| 66 | PRPF4B | 3208 5295 | 11612 | -0.041 | 0.1163 | No | ||
| 67 | SNRPE | 9843 | 11630 | -0.041 | 0.1162 | No | ||
| 68 | DHX15 | 8842 | 11985 | -0.049 | 0.0979 | No | ||
| 69 | POLR2A | 5394 | 12113 | -0.052 | 0.0920 | No | ||
| 70 | DDX20 | 15213 | 12761 | -0.070 | 0.0584 | No | ||
| 71 | XRN2 | 2936 6302 2905 | 13216 | -0.086 | 0.0354 | No | ||
| 72 | PTBP2 | 7074 | 13221 | -0.086 | 0.0368 | No | ||
| 73 | SNRPA1 | 1373 18216 | 13242 | -0.087 | 0.0373 | No | ||
| 74 | LSM2 | 23266 | 13660 | -0.107 | 0.0167 | No | ||
| 75 | HNRPL | 18315 | 13864 | -0.119 | 0.0080 | No | ||
| 76 | RNMT | 7501 | 14503 | -0.169 | -0.0234 | No | ||
| 77 | CPSF2 | 21182 | 14637 | -0.184 | -0.0272 | No | ||
| 78 | DNAJC8 | 12712 | 15198 | -0.255 | -0.0528 | No | ||
| 79 | SF3A2 | 19938 | 15228 | -0.260 | -0.0496 | No | ||
| 80 | METTL3 | 21841 | 15575 | -0.326 | -0.0623 | No | ||
| 81 | SF3B5 | 7256 | 15576 | -0.326 | -0.0563 | No | ||
| 82 | SRPK2 | 5513 | 15688 | -0.354 | -0.0558 | No | ||
| 83 | HNRPC | 9102 4859 | 15731 | -0.362 | -0.0514 | No | ||
| 84 | SNRPA | 7007 11992 7008 | 15807 | -0.379 | -0.0485 | No | ||
| 85 | FUSIP1 | 4715 16036 | 15900 | -0.403 | -0.0461 | No | ||
| 86 | SF3B1 | 13954 | 15970 | -0.423 | -0.0421 | No | ||
| 87 | SFRS7 | 22889 | 16062 | -0.447 | -0.0388 | No | ||
| 88 | TXNL4A | 6567 11329 | 16106 | -0.458 | -0.0327 | No | ||
| 89 | CSTF2 | 24257 | 16129 | -0.467 | -0.0254 | No | ||
| 90 | NCBP2 | 12643 | 16223 | -0.492 | -0.0214 | No | ||
| 91 | SNRPD1 | 23622 | 16738 | -0.693 | -0.0364 | No | ||
| 92 | SNRPG | 12622 | 16958 | -0.803 | -0.0335 | No | ||
| 93 | CPSF4 | 16627 | 17194 | -0.918 | -0.0294 | No | ||
| 94 | HNRPH1 | 12222 1486 1220 7202 7201 | 17386 | -1.005 | -0.0213 | No | ||
| 95 | CSTF3 | 10449 6002 | 17511 | -1.069 | -0.0084 | No | ||
| 96 | CSTF1 | 12515 | 17566 | -1.105 | 0.0089 | No | ||
| 97 | SNRPD2 | 8412 | 17680 | -1.170 | 0.0243 | No | ||
| 98 | LSM7 | 12286 | 18026 | -1.430 | 0.0319 | No |

