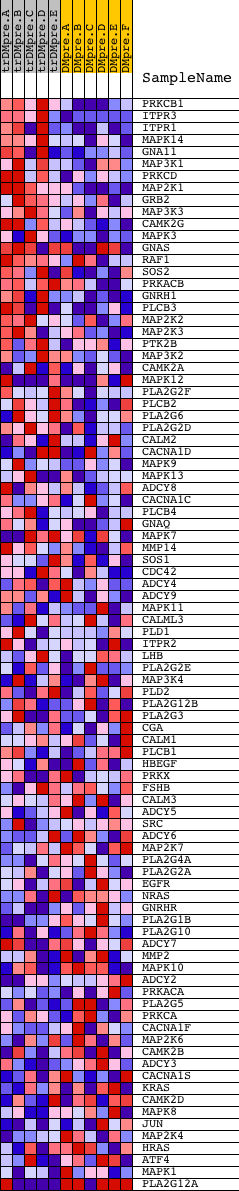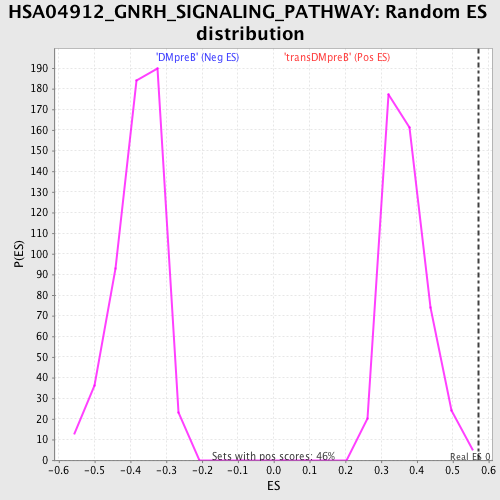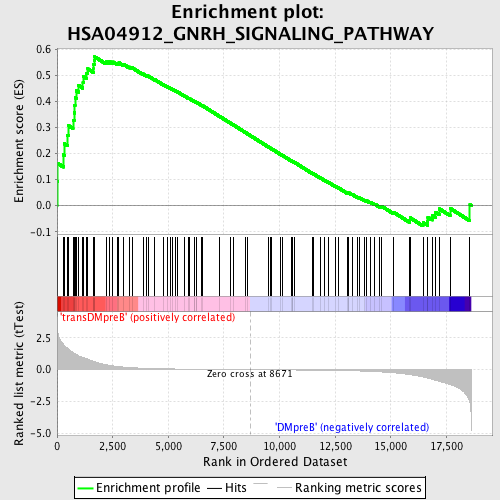
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | HSA04912_GNRH_SIGNALING_PATHWAY |
| Enrichment Score (ES) | 0.57063955 |
| Normalized Enrichment Score (NES) | 1.5468112 |
| Nominal p-value | 0.0021691974 |
| FDR q-value | 0.3281005 |
| FWER p-Value | 0.944 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PRKCB1 | 1693 9574 | 2 | 3.865 | 0.0928 | Yes | ||
| 2 | ITPR3 | 9195 | 31 | 2.936 | 0.1618 | Yes | ||
| 3 | ITPR1 | 17341 | 285 | 1.959 | 0.1952 | Yes | ||
| 4 | MAPK14 | 23313 | 334 | 1.857 | 0.2373 | Yes | ||
| 5 | GNA11 | 3300 3345 19670 | 465 | 1.674 | 0.2705 | Yes | ||
| 6 | MAP3K1 | 21348 | 512 | 1.620 | 0.3069 | Yes | ||
| 7 | PRKCD | 21897 | 716 | 1.338 | 0.3281 | Yes | ||
| 8 | MAP2K1 | 19082 | 767 | 1.285 | 0.3563 | Yes | ||
| 9 | GRB2 | 20149 | 792 | 1.266 | 0.3854 | Yes | ||
| 10 | MAP3K3 | 20626 | 808 | 1.254 | 0.4147 | Yes | ||
| 11 | CAMK2G | 21905 | 866 | 1.191 | 0.4403 | Yes | ||
| 12 | MAPK3 | 6458 11170 | 961 | 1.090 | 0.4614 | Yes | ||
| 13 | GNAS | 9025 2963 2752 | 1162 | 0.961 | 0.4737 | Yes | ||
| 14 | RAF1 | 17035 | 1186 | 0.942 | 0.4951 | Yes | ||
| 15 | SOS2 | 21049 | 1331 | 0.845 | 0.5076 | Yes | ||
| 16 | PRKACB | 15140 | 1381 | 0.813 | 0.5246 | Yes | ||
| 17 | GNRH1 | 125 | 1630 | 0.664 | 0.5271 | Yes | ||
| 18 | PLCB3 | 23799 | 1635 | 0.661 | 0.5428 | Yes | ||
| 19 | MAP2K2 | 19933 | 1669 | 0.648 | 0.5566 | Yes | ||
| 20 | MAP2K3 | 20856 | 1692 | 0.634 | 0.5706 | Yes | ||
| 21 | PTK2B | 21776 | 2197 | 0.385 | 0.5527 | No | ||
| 22 | MAP3K2 | 11165 | 2345 | 0.340 | 0.5529 | No | ||
| 23 | CAMK2A | 2024 23541 1980 | 2511 | 0.290 | 0.5510 | No | ||
| 24 | MAPK12 | 2182 2215 22164 | 2721 | 0.246 | 0.5456 | No | ||
| 25 | PLA2G2F | 15700 | 2780 | 0.236 | 0.5481 | No | ||
| 26 | PLCB2 | 5262 | 2988 | 0.201 | 0.5418 | No | ||
| 27 | PLA2G6 | 22209 | 3248 | 0.165 | 0.5318 | No | ||
| 28 | PLA2G2D | 16018 | 3366 | 0.151 | 0.5291 | No | ||
| 29 | CALM2 | 8681 | 3860 | 0.110 | 0.5051 | No | ||
| 30 | CACNA1D | 4464 8675 | 4027 | 0.100 | 0.4986 | No | ||
| 31 | MAPK9 | 1233 20903 1383 | 4094 | 0.096 | 0.4973 | No | ||
| 32 | MAPK13 | 23314 | 4397 | 0.080 | 0.4829 | No | ||
| 33 | ADCY8 | 22281 | 4797 | 0.065 | 0.4630 | No | ||
| 34 | CACNA1C | 1158 1166 24464 4463 8674 | 4954 | 0.060 | 0.4560 | No | ||
| 35 | PLCB4 | 9586 | 5094 | 0.056 | 0.4498 | No | ||
| 36 | GNAQ | 4786 23909 3685 | 5182 | 0.054 | 0.4464 | No | ||
| 37 | MAPK7 | 1381 20414 | 5307 | 0.050 | 0.4409 | No | ||
| 38 | MMP14 | 22020 | 5398 | 0.048 | 0.4372 | No | ||
| 39 | SOS1 | 5476 | 5727 | 0.041 | 0.4205 | No | ||
| 40 | CDC42 | 4503 8722 4504 2465 | 5895 | 0.038 | 0.4124 | No | ||
| 41 | ADCY4 | 2980 21818 | 5954 | 0.036 | 0.4102 | No | ||
| 42 | ADCY9 | 22680 | 6166 | 0.033 | 0.3996 | No | ||
| 43 | MAPK11 | 2264 9618 22163 | 6170 | 0.033 | 0.4002 | No | ||
| 44 | CALML3 | 21553 | 6249 | 0.032 | 0.3968 | No | ||
| 45 | PLD1 | 15624 | 6254 | 0.032 | 0.3973 | No | ||
| 46 | ITPR2 | 9194 | 6505 | 0.028 | 0.3845 | No | ||
| 47 | LHB | 4993 | 6551 | 0.027 | 0.3827 | No | ||
| 48 | PLA2G2E | 16016 | 7318 | 0.016 | 0.3417 | No | ||
| 49 | MAP3K4 | 23126 | 7801 | 0.010 | 0.3159 | No | ||
| 50 | PLD2 | 20803 | 7913 | 0.009 | 0.3102 | No | ||
| 51 | PLA2G12B | 20016 | 8454 | 0.003 | 0.2811 | No | ||
| 52 | PLA2G3 | 20968 | 8577 | 0.001 | 0.2745 | No | ||
| 53 | CGA | 16246 | 9520 | -0.011 | 0.2239 | No | ||
| 54 | CALM1 | 21184 | 9606 | -0.012 | 0.2196 | No | ||
| 55 | PLCB1 | 14832 2821 | 9656 | -0.012 | 0.2173 | No | ||
| 56 | HBEGF | 23457 | 10038 | -0.017 | 0.1971 | No | ||
| 57 | PRKX | 9619 5290 | 10149 | -0.019 | 0.1916 | No | ||
| 58 | FSHB | 14500 | 10523 | -0.024 | 0.1721 | No | ||
| 59 | CALM3 | 8682 | 10585 | -0.025 | 0.1694 | No | ||
| 60 | ADCY5 | 10312 | 10596 | -0.025 | 0.1694 | No | ||
| 61 | SRC | 5507 | 10688 | -0.026 | 0.1652 | No | ||
| 62 | ADCY6 | 22139 2283 8551 | 11490 | -0.039 | 0.1229 | No | ||
| 63 | MAP2K7 | 6453 | 11507 | -0.039 | 0.1229 | No | ||
| 64 | PLA2G4A | 13809 | 11835 | -0.046 | 0.1064 | No | ||
| 65 | PLA2G2A | 16017 | 12015 | -0.049 | 0.0979 | No | ||
| 66 | EGFR | 1329 20944 | 12205 | -0.054 | 0.0890 | No | ||
| 67 | NRAS | 5191 | 12495 | -0.061 | 0.0749 | No | ||
| 68 | GNRHR | 4791 | 12632 | -0.066 | 0.0691 | No | ||
| 69 | PLA2G1B | 9581 | 13039 | -0.079 | 0.0491 | No | ||
| 70 | PLA2G10 | 22658 | 13105 | -0.082 | 0.0475 | No | ||
| 71 | ADCY7 | 8552 448 | 13118 | -0.082 | 0.0489 | No | ||
| 72 | MMP2 | 18525 | 13265 | -0.088 | 0.0431 | No | ||
| 73 | MAPK10 | 11169 | 13511 | -0.099 | 0.0323 | No | ||
| 74 | ADCY2 | 21412 | 13602 | -0.104 | 0.0299 | No | ||
| 75 | PRKACA | 18549 3844 | 13814 | -0.116 | 0.0213 | No | ||
| 76 | PLA2G5 | 9583 | 13892 | -0.120 | 0.0201 | No | ||
| 77 | PRKCA | 20174 | 14071 | -0.131 | 0.0136 | No | ||
| 78 | CACNA1F | 12054 24393 | 14271 | -0.147 | 0.0064 | No | ||
| 79 | MAP2K6 | 20614 1414 | 14512 | -0.170 | -0.0025 | No | ||
| 80 | CAMK2B | 20536 | 14588 | -0.178 | -0.0022 | No | ||
| 81 | ADCY3 | 21329 894 | 15106 | -0.241 | -0.0244 | No | ||
| 82 | CACNA1S | 14111 | 15856 | -0.391 | -0.0554 | No | ||
| 83 | KRAS | 9247 | 15861 | -0.392 | -0.0462 | No | ||
| 84 | CAMK2D | 4232 | 16448 | -0.567 | -0.0642 | No | ||
| 85 | MAPK8 | 6459 | 16644 | -0.646 | -0.0592 | No | ||
| 86 | JUN | 15832 | 16667 | -0.656 | -0.0446 | No | ||
| 87 | MAP2K4 | 20405 | 16860 | -0.753 | -0.0369 | No | ||
| 88 | HRAS | 4868 | 17006 | -0.832 | -0.0247 | No | ||
| 89 | ATF4 | 22417 2239 | 17185 | -0.914 | -0.0124 | No | ||
| 90 | MAPK1 | 1642 11167 | 17667 | -1.160 | -0.0104 | No | ||
| 91 | PLA2G12A | 1782 7281 1839 | 18556 | -2.564 | 0.0032 | No |