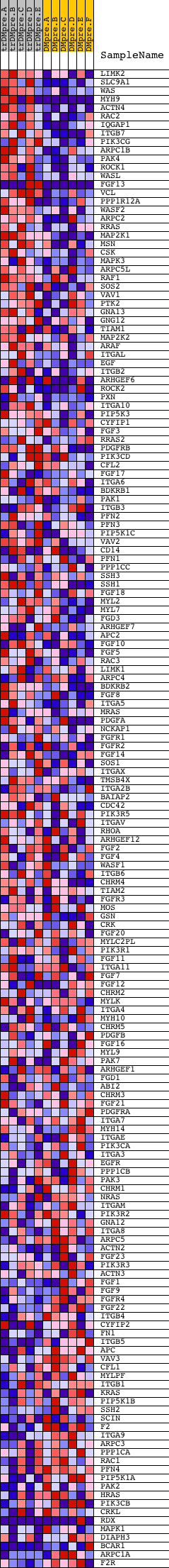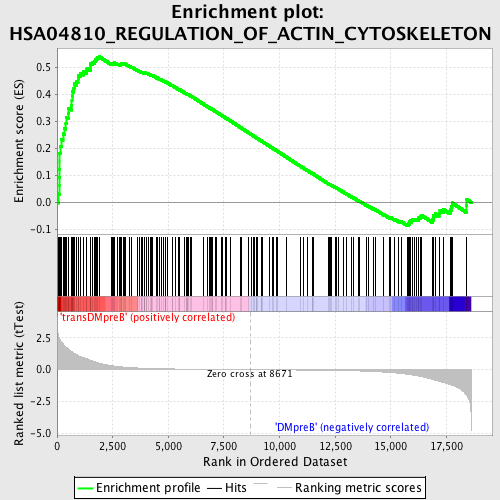
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | HSA04810_REGULATION_OF_ACTIN_CYTOSKELETON |
| Enrichment Score (ES) | 0.5378721 |
| Normalized Enrichment Score (NES) | 1.5866159 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.27389315 |
| FWER p-Value | 0.829 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | LIMK2 | 9278 1296 | 47 | 2.806 | 0.0326 | Yes | ||
| 2 | SLC9A1 | 16053 | 96 | 2.468 | 0.0609 | Yes | ||
| 3 | WAS | 24193 | 99 | 2.439 | 0.0913 | Yes | ||
| 4 | MYH9 | 2252 2244 | 109 | 2.414 | 0.1210 | Yes | ||
| 5 | ACTN4 | 17905 1798 1983 | 116 | 2.388 | 0.1506 | Yes | ||
| 6 | RAC2 | 22217 | 117 | 2.387 | 0.1804 | Yes | ||
| 7 | IQGAP1 | 6619 | 136 | 2.288 | 0.2081 | Yes | ||
| 8 | ITGB7 | 22112 | 191 | 2.180 | 0.2324 | Yes | ||
| 9 | PIK3CG | 6635 | 265 | 2.027 | 0.2538 | Yes | ||
| 10 | ARPC1B | 3512 8628 | 325 | 1.871 | 0.2741 | Yes | ||
| 11 | PAK4 | 17909 | 396 | 1.777 | 0.2925 | Yes | ||
| 12 | ROCK1 | 5386 | 409 | 1.746 | 0.3137 | Yes | ||
| 13 | WASL | 17203 | 501 | 1.629 | 0.3292 | Yes | ||
| 14 | FGF13 | 4720 8963 2627 | 518 | 1.613 | 0.3485 | Yes | ||
| 15 | VCL | 22083 | 654 | 1.422 | 0.3589 | Yes | ||
| 16 | PPP1R12A | 3354 19886 | 668 | 1.402 | 0.3758 | Yes | ||
| 17 | WASF2 | 6326 | 671 | 1.401 | 0.3932 | Yes | ||
| 18 | ARPC2 | 14225 | 677 | 1.393 | 0.4104 | Yes | ||
| 19 | RRAS | 18256 | 752 | 1.300 | 0.4226 | Yes | ||
| 20 | MAP2K1 | 19082 | 767 | 1.285 | 0.4380 | Yes | ||
| 21 | MSN | 5122 | 879 | 1.181 | 0.4467 | Yes | ||
| 22 | CSK | 8805 | 940 | 1.117 | 0.4574 | Yes | ||
| 23 | MAPK3 | 6458 11170 | 961 | 1.090 | 0.4700 | Yes | ||
| 24 | ARPC5L | 15017 | 1060 | 1.013 | 0.4774 | Yes | ||
| 25 | RAF1 | 17035 | 1186 | 0.942 | 0.4824 | Yes | ||
| 26 | SOS2 | 21049 | 1331 | 0.845 | 0.4851 | Yes | ||
| 27 | VAV1 | 23173 | 1340 | 0.841 | 0.4952 | Yes | ||
| 28 | PTK2 | 22271 | 1498 | 0.741 | 0.4960 | Yes | ||
| 29 | GNA13 | 20617 | 1502 | 0.738 | 0.5051 | Yes | ||
| 30 | GNG12 | 4789 | 1518 | 0.728 | 0.5134 | Yes | ||
| 31 | TIAM1 | 22547 | 1578 | 0.694 | 0.5188 | Yes | ||
| 32 | MAP2K2 | 19933 | 1669 | 0.648 | 0.5221 | Yes | ||
| 33 | ARAF | 24367 | 1729 | 0.613 | 0.5265 | Yes | ||
| 34 | ITGAL | 9187 | 1788 | 0.569 | 0.5305 | Yes | ||
| 35 | EGF | 15169 | 1801 | 0.561 | 0.5369 | Yes | ||
| 36 | ITGB2 | 19978 | 1901 | 0.507 | 0.5379 | Yes | ||
| 37 | ARHGEF6 | 13297 13104 | 2444 | 0.307 | 0.5123 | No | ||
| 38 | ROCK2 | 21309 5387 | 2496 | 0.295 | 0.5132 | No | ||
| 39 | PXN | 5339 3573 | 2545 | 0.281 | 0.5142 | No | ||
| 40 | ITGA10 | 15493 | 2557 | 0.279 | 0.5171 | No | ||
| 41 | PIP5K3 | 14234 | 2729 | 0.245 | 0.5108 | No | ||
| 42 | CYFIP1 | 9814 | 2792 | 0.234 | 0.5104 | No | ||
| 43 | FGF3 | 17997 | 2866 | 0.222 | 0.5092 | No | ||
| 44 | RRAS2 | 12431 | 2893 | 0.217 | 0.5105 | No | ||
| 45 | PDGFRB | 23539 | 2909 | 0.214 | 0.5124 | No | ||
| 46 | PIK3CD | 9563 | 2973 | 0.204 | 0.5115 | No | ||
| 47 | CFL2 | 2155 21069 | 3023 | 0.197 | 0.5113 | No | ||
| 48 | FGF17 | 21757 | 3061 | 0.193 | 0.5118 | No | ||
| 49 | ITGA6 | 4930 | 3269 | 0.163 | 0.5026 | No | ||
| 50 | BDKRB1 | 19418 | 3347 | 0.153 | 0.5003 | No | ||
| 51 | PAK1 | 9527 | 3601 | 0.130 | 0.4882 | No | ||
| 52 | ITGB3 | 20631 | 3713 | 0.121 | 0.4837 | No | ||
| 53 | PFN2 | 15337 1818 | 3777 | 0.116 | 0.4817 | No | ||
| 54 | PFN3 | 7968 | 3812 | 0.114 | 0.4813 | No | ||
| 55 | PIP5K1C | 5250 19929 | 3843 | 0.112 | 0.4811 | No | ||
| 56 | VAV2 | 5848 2670 | 3906 | 0.108 | 0.4791 | No | ||
| 57 | CD14 | 23452 | 3922 | 0.107 | 0.4796 | No | ||
| 58 | PFN1 | 9555 | 3931 | 0.106 | 0.4805 | No | ||
| 59 | PPP1CC | 9609 5283 | 4016 | 0.101 | 0.4772 | No | ||
| 60 | SSH3 | 3709 10890 | 4039 | 0.100 | 0.4773 | No | ||
| 61 | SSH1 | 10540 | 4114 | 0.095 | 0.4744 | No | ||
| 62 | FGF18 | 4721 1351 | 4204 | 0.089 | 0.4707 | No | ||
| 63 | MYL2 | 16713 | 4236 | 0.087 | 0.4701 | No | ||
| 64 | MYL7 | 9437 | 4262 | 0.086 | 0.4699 | No | ||
| 65 | FGD3 | 11399 | 4280 | 0.085 | 0.4700 | No | ||
| 66 | ARHGEF7 | 3823 | 4485 | 0.076 | 0.4599 | No | ||
| 67 | APC2 | 10704 | 4489 | 0.076 | 0.4607 | No | ||
| 68 | FGF10 | 4719 8962 | 4595 | 0.072 | 0.4559 | No | ||
| 69 | FGF5 | 16785 | 4618 | 0.071 | 0.4556 | No | ||
| 70 | RAC3 | 20561 | 4683 | 0.069 | 0.4530 | No | ||
| 71 | LIMK1 | 16350 | 4789 | 0.065 | 0.4481 | No | ||
| 72 | ARPC4 | 12642 | 4857 | 0.063 | 0.4453 | No | ||
| 73 | BDKRB2 | 21163 | 4876 | 0.063 | 0.4451 | No | ||
| 74 | FGF8 | 23656 | 4879 | 0.063 | 0.4458 | No | ||
| 75 | ITGA5 | 22105 | 4957 | 0.060 | 0.4423 | No | ||
| 76 | MRAS | 9409 | 5166 | 0.054 | 0.4317 | No | ||
| 77 | PDGFA | 5237 | 5187 | 0.054 | 0.4313 | No | ||
| 78 | NCKAP1 | 6947 | 5339 | 0.050 | 0.4238 | No | ||
| 79 | FGFR1 | 3789 8968 | 5469 | 0.047 | 0.4174 | No | ||
| 80 | FGFR2 | 1917 4722 1119 | 5517 | 0.046 | 0.4154 | No | ||
| 81 | FGF14 | 21716 3005 3463 | 5520 | 0.046 | 0.4158 | No | ||
| 82 | SOS1 | 5476 | 5727 | 0.041 | 0.4052 | No | ||
| 83 | ITGAX | 18058 | 5813 | 0.040 | 0.4011 | No | ||
| 84 | TMSB4X | 24005 | 5836 | 0.039 | 0.4004 | No | ||
| 85 | ITGA2B | 9186 | 5847 | 0.039 | 0.4003 | No | ||
| 86 | BAIAP2 | 449 4236 | 5879 | 0.038 | 0.3991 | No | ||
| 87 | CDC42 | 4503 8722 4504 2465 | 5895 | 0.038 | 0.3988 | No | ||
| 88 | PIK3R5 | 11507 | 5998 | 0.036 | 0.3937 | No | ||
| 89 | ITGAV | 4931 | 6021 | 0.036 | 0.3929 | No | ||
| 90 | RHOA | 8624 4409 4410 | 6560 | 0.027 | 0.3641 | No | ||
| 91 | ARHGEF12 | 19156 | 6766 | 0.024 | 0.3533 | No | ||
| 92 | FGF2 | 15608 | 6855 | 0.022 | 0.3488 | No | ||
| 93 | FGF4 | 8964 | 6883 | 0.022 | 0.3476 | No | ||
| 94 | WASF1 | 13514 | 6956 | 0.021 | 0.3439 | No | ||
| 95 | ITGB6 | 14581 2809 | 6994 | 0.020 | 0.3422 | No | ||
| 96 | CHRM4 | 14945 | 6996 | 0.020 | 0.3424 | No | ||
| 97 | TIAM2 | 10759 | 7123 | 0.019 | 0.3358 | No | ||
| 98 | FGFR3 | 8969 3566 | 7141 | 0.018 | 0.3351 | No | ||
| 99 | MOS | 16254 | 7388 | 0.015 | 0.3219 | No | ||
| 100 | GSN | 2784 | 7436 | 0.014 | 0.3196 | No | ||
| 101 | CRK | 4559 1249 | 7549 | 0.013 | 0.3137 | No | ||
| 102 | FGF20 | 18892 | 7588 | 0.013 | 0.3118 | No | ||
| 103 | MYLC2PL | 16670 | 7624 | 0.012 | 0.3100 | No | ||
| 104 | PIK3R1 | 3170 | 7780 | 0.010 | 0.3017 | No | ||
| 105 | FGF11 | 20383 | 8220 | 0.005 | 0.2780 | No | ||
| 106 | ITGA11 | 6677 11449 | 8300 | 0.004 | 0.2738 | No | ||
| 107 | FGF7 | 8965 | 8584 | 0.001 | 0.2584 | No | ||
| 108 | FGF12 | 1723 22621 | 8735 | -0.001 | 0.2503 | No | ||
| 109 | CHRM2 | 10840 | 8814 | -0.002 | 0.2461 | No | ||
| 110 | MYLK | 22778 4213 | 8871 | -0.002 | 0.2431 | No | ||
| 111 | ITGA4 | 4929 | 8961 | -0.003 | 0.2383 | No | ||
| 112 | MYH10 | 8077 | 8967 | -0.003 | 0.2381 | No | ||
| 113 | CHRM5 | 10038 | 8995 | -0.004 | 0.2367 | No | ||
| 114 | PDGFB | 22199 2311 | 9172 | -0.006 | 0.2272 | No | ||
| 115 | FGF16 | 24270 | 9187 | -0.006 | 0.2265 | No | ||
| 116 | MYL9 | 14767 2659 | 9251 | -0.007 | 0.2232 | No | ||
| 117 | PAK7 | 14417 | 9538 | -0.011 | 0.2078 | No | ||
| 118 | ARHGEF1 | 1952 18343 3996 | 9677 | -0.013 | 0.2005 | No | ||
| 119 | FGD1 | 24228 | 9726 | -0.013 | 0.1980 | No | ||
| 120 | ABI2 | 4037 6830 | 9877 | -0.015 | 0.1901 | No | ||
| 121 | CHRM3 | 21547 | 9913 | -0.016 | 0.1884 | No | ||
| 122 | FGF21 | 17830 | 10314 | -0.021 | 0.1670 | No | ||
| 123 | PDGFRA | 16824 | 10927 | -0.030 | 0.1342 | No | ||
| 124 | ITGA7 | 19835 | 11089 | -0.032 | 0.1258 | No | ||
| 125 | MYH14 | 17845 | 11234 | -0.034 | 0.1185 | No | ||
| 126 | ITGAE | 20791 1481 1397 1380 1278 1238 1359 | 11462 | -0.038 | 0.1066 | No | ||
| 127 | PIK3CA | 9562 | 11493 | -0.039 | 0.1055 | No | ||
| 128 | ITGA3 | 20284 | 11505 | -0.039 | 0.1054 | No | ||
| 129 | EGFR | 1329 20944 | 12205 | -0.054 | 0.0681 | No | ||
| 130 | PPP1CB | 5282 3529 3504 | 12239 | -0.055 | 0.0670 | No | ||
| 131 | PAK3 | 9528 | 12282 | -0.056 | 0.0655 | No | ||
| 132 | CHRM1 | 23943 | 12322 | -0.057 | 0.0641 | No | ||
| 133 | NRAS | 5191 | 12495 | -0.061 | 0.0555 | No | ||
| 134 | ITGAM | 9188 | 12558 | -0.063 | 0.0529 | No | ||
| 135 | PIK3R2 | 18850 | 12662 | -0.067 | 0.0482 | No | ||
| 136 | GNA12 | 9023 | 12854 | -0.073 | 0.0387 | No | ||
| 137 | ITGA8 | 14685 | 12999 | -0.078 | 0.0319 | No | ||
| 138 | ARPC5 | 12580 7487 | 13211 | -0.086 | 0.0215 | No | ||
| 139 | ACTN2 | 21545 | 13310 | -0.090 | 0.0173 | No | ||
| 140 | FGF23 | 17275 | 13546 | -0.101 | 0.0059 | No | ||
| 141 | PIK3R3 | 5248 | 13606 | -0.105 | 0.0040 | No | ||
| 142 | ACTN3 | 23967 8540 | 13926 | -0.122 | -0.0118 | No | ||
| 143 | FGF1 | 1994 23447 | 14006 | -0.128 | -0.0145 | No | ||
| 144 | FGF9 | 8966 | 14217 | -0.143 | -0.0241 | No | ||
| 145 | FGFR4 | 3226 | 14308 | -0.151 | -0.0271 | No | ||
| 146 | FGF22 | 12468 7385 | 14687 | -0.190 | -0.0452 | No | ||
| 147 | ITGB4 | 9669 159 | 14952 | -0.222 | -0.0567 | No | ||
| 148 | CYFIP2 | 20485 8045 | 14966 | -0.224 | -0.0546 | No | ||
| 149 | FN1 | 4091 4094 971 13929 | 15177 | -0.253 | -0.0629 | No | ||
| 150 | ITGB5 | 4932 | 15363 | -0.283 | -0.0694 | No | ||
| 151 | APC | 4396 2022 | 15464 | -0.302 | -0.0710 | No | ||
| 152 | VAV3 | 1774 1848 15443 | 15768 | -0.369 | -0.0828 | No | ||
| 153 | CFL1 | 4516 | 15803 | -0.378 | -0.0799 | No | ||
| 154 | MYLPF | 18069 | 15833 | -0.385 | -0.0767 | No | ||
| 155 | ITGB1 | 3872 18411 | 15843 | -0.387 | -0.0723 | No | ||
| 156 | KRAS | 9247 | 15861 | -0.392 | -0.0683 | No | ||
| 157 | PIP5K1B | 5251 1841 | 15951 | -0.416 | -0.0680 | No | ||
| 158 | SSH2 | 6202 | 15972 | -0.423 | -0.0637 | No | ||
| 159 | SCIN | 21079 | 16068 | -0.450 | -0.0633 | No | ||
| 160 | F2 | 14524 | 16153 | -0.473 | -0.0619 | No | ||
| 161 | ITGA9 | 19280 | 16241 | -0.496 | -0.0604 | No | ||
| 162 | ARPC3 | 12109 | 16258 | -0.501 | -0.0550 | No | ||
| 163 | PPP1CA | 9608 3756 914 909 | 16336 | -0.531 | -0.0525 | No | ||
| 164 | RAC1 | 16302 | 16377 | -0.547 | -0.0479 | No | ||
| 165 | PFN4 | 11798 | 16863 | -0.753 | -0.0647 | No | ||
| 166 | PIP5K1A | 23708 | 16908 | -0.776 | -0.0574 | No | ||
| 167 | PAK2 | 10310 5875 | 16924 | -0.783 | -0.0484 | No | ||
| 168 | HRAS | 4868 | 17006 | -0.832 | -0.0424 | No | ||
| 169 | PIK3CB | 19030 | 17167 | -0.904 | -0.0398 | No | ||
| 170 | CRKL | 4560 | 17208 | -0.926 | -0.0304 | No | ||
| 171 | RDX | 9712 5371 | 17373 | -1.002 | -0.0267 | No | ||
| 172 | MAPK1 | 1642 11167 | 17667 | -1.160 | -0.0281 | No | ||
| 173 | DIAPH3 | 7117 | 17711 | -1.189 | -0.0155 | No | ||
| 174 | BCAR1 | 18741 | 17751 | -1.220 | -0.0024 | No | ||
| 175 | ARPC1A | 3541 16628 3519 | 18395 | -1.949 | -0.0129 | No | ||
| 176 | F2R | 21386 | 18408 | -1.981 | 0.0113 | No |