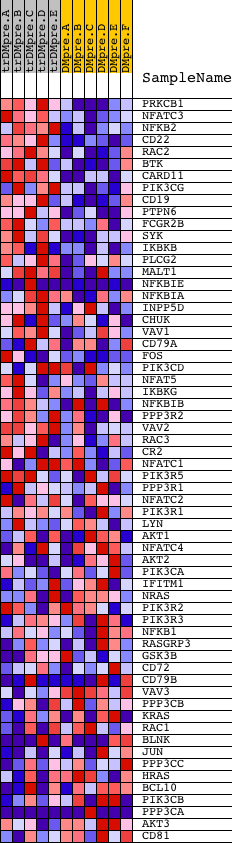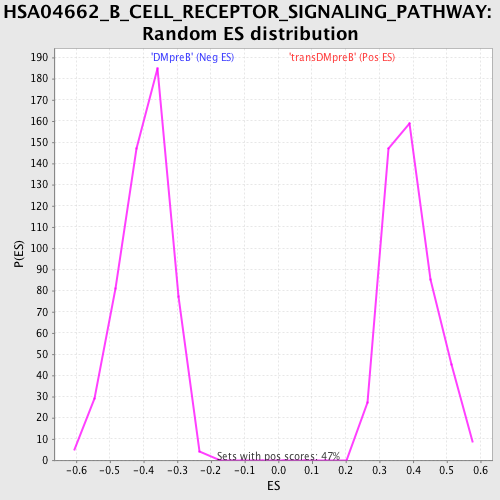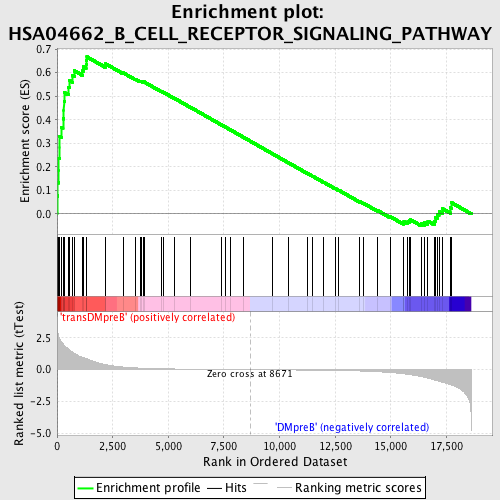
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | HSA04662_B_CELL_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | 0.6670168 |
| Normalized Enrichment Score (NES) | 1.7108221 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.13667168 |
| FWER p-Value | 0.175 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PRKCB1 | 1693 9574 | 2 | 3.865 | 0.0771 | Yes | ||
| 2 | NFATC3 | 5169 | 38 | 2.872 | 0.1326 | Yes | ||
| 3 | NFKB2 | 23810 | 65 | 2.638 | 0.1839 | Yes | ||
| 4 | CD22 | 17881 | 70 | 2.621 | 0.2360 | Yes | ||
| 5 | RAC2 | 22217 | 117 | 2.387 | 0.2812 | Yes | ||
| 6 | BTK | 24061 | 118 | 2.378 | 0.3287 | Yes | ||
| 7 | CARD11 | 8436 | 186 | 2.186 | 0.3688 | Yes | ||
| 8 | PIK3CG | 6635 | 265 | 2.027 | 0.4051 | Yes | ||
| 9 | CD19 | 17640 | 306 | 1.909 | 0.4410 | Yes | ||
| 10 | PTPN6 | 17002 | 309 | 1.905 | 0.4790 | Yes | ||
| 11 | FCGR2B | 8959 | 336 | 1.855 | 0.5147 | Yes | ||
| 12 | SYK | 21636 | 513 | 1.619 | 0.5375 | Yes | ||
| 13 | IKBKB | 4907 | 558 | 1.550 | 0.5661 | Yes | ||
| 14 | PLCG2 | 18453 | 681 | 1.392 | 0.5873 | Yes | ||
| 15 | MALT1 | 6274 | 775 | 1.279 | 0.6079 | Yes | ||
| 16 | NFKBIE | 23225 1556 | 1147 | 0.973 | 0.6073 | Yes | ||
| 17 | NFKBIA | 21065 | 1178 | 0.947 | 0.6246 | Yes | ||
| 18 | INPP5D | 14198 | 1301 | 0.857 | 0.6352 | Yes | ||
| 19 | CHUK | 23665 | 1310 | 0.853 | 0.6518 | Yes | ||
| 20 | VAV1 | 23173 | 1340 | 0.841 | 0.6670 | Yes | ||
| 21 | CD79A | 18342 | 2161 | 0.395 | 0.6307 | No | ||
| 22 | FOS | 21202 | 2168 | 0.392 | 0.6382 | No | ||
| 23 | PIK3CD | 9563 | 2973 | 0.204 | 0.5990 | No | ||
| 24 | NFAT5 | 3921 7037 12036 | 3519 | 0.137 | 0.5723 | No | ||
| 25 | IKBKG | 2570 2562 4908 | 3736 | 0.119 | 0.5631 | No | ||
| 26 | NFKBIB | 17906 | 3771 | 0.117 | 0.5636 | No | ||
| 27 | PPP3R2 | 9612 | 3876 | 0.109 | 0.5601 | No | ||
| 28 | VAV2 | 5848 2670 | 3906 | 0.108 | 0.5607 | No | ||
| 29 | RAC3 | 20561 | 4683 | 0.069 | 0.5203 | No | ||
| 30 | CR2 | 3942 13707 | 4795 | 0.065 | 0.5156 | No | ||
| 31 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 5266 | 0.052 | 0.4913 | No | ||
| 32 | PIK3R5 | 11507 | 5998 | 0.036 | 0.4526 | No | ||
| 33 | PPP3R1 | 9611 489 | 7403 | 0.015 | 0.3773 | No | ||
| 34 | NFATC2 | 5168 2866 | 7581 | 0.013 | 0.3680 | No | ||
| 35 | PIK3R1 | 3170 | 7780 | 0.010 | 0.3575 | No | ||
| 36 | LYN | 16281 | 8372 | 0.004 | 0.3257 | No | ||
| 37 | AKT1 | 8568 | 9662 | -0.013 | 0.2565 | No | ||
| 38 | NFATC4 | 22002 | 10388 | -0.022 | 0.2179 | No | ||
| 39 | AKT2 | 4365 4366 | 11247 | -0.035 | 0.1723 | No | ||
| 40 | PIK3CA | 9562 | 11493 | -0.039 | 0.1599 | No | ||
| 41 | IFITM1 | 18017 7578 12724 | 11969 | -0.048 | 0.1352 | No | ||
| 42 | NRAS | 5191 | 12495 | -0.061 | 0.1082 | No | ||
| 43 | PIK3R2 | 18850 | 12662 | -0.067 | 0.1006 | No | ||
| 44 | PIK3R3 | 5248 | 13606 | -0.105 | 0.0518 | No | ||
| 45 | NFKB1 | 15160 | 13771 | -0.113 | 0.0453 | No | ||
| 46 | RASGRP3 | 10767 | 14421 | -0.162 | 0.0135 | No | ||
| 47 | GSK3B | 22761 | 14988 | -0.227 | -0.0125 | No | ||
| 48 | CD72 | 8718 | 15559 | -0.322 | -0.0367 | No | ||
| 49 | CD79B | 20185 1309 | 15588 | -0.328 | -0.0317 | No | ||
| 50 | VAV3 | 1774 1848 15443 | 15768 | -0.369 | -0.0340 | No | ||
| 51 | PPP3CB | 5285 | 15826 | -0.383 | -0.0294 | No | ||
| 52 | KRAS | 9247 | 15861 | -0.392 | -0.0234 | No | ||
| 53 | RAC1 | 16302 | 16377 | -0.547 | -0.0402 | No | ||
| 54 | BLNK | 23681 3691 | 16514 | -0.592 | -0.0357 | No | ||
| 55 | JUN | 15832 | 16667 | -0.656 | -0.0308 | No | ||
| 56 | PPP3CC | 21763 | 16969 | -0.809 | -0.0308 | No | ||
| 57 | HRAS | 4868 | 17006 | -0.832 | -0.0162 | No | ||
| 58 | BCL10 | 15397 | 17084 | -0.870 | -0.0029 | No | ||
| 59 | PIK3CB | 19030 | 17167 | -0.904 | 0.0107 | No | ||
| 60 | PPP3CA | 1863 5284 | 17331 | -0.993 | 0.0218 | No | ||
| 61 | AKT3 | 13739 982 | 17683 | -1.172 | 0.0262 | No | ||
| 62 | CD81 | 8719 | 17724 | -1.201 | 0.0481 | No |