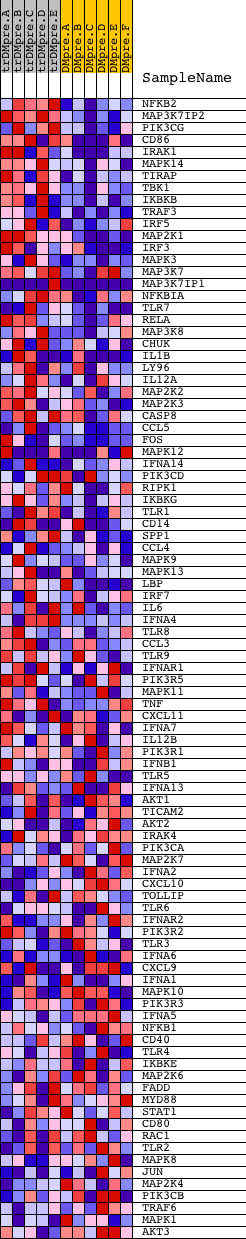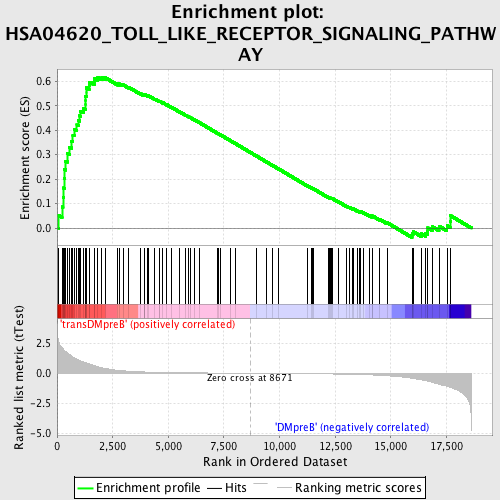
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | HSA04620_TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | 0.6151787 |
| Normalized Enrichment Score (NES) | 1.6684479 |
| Nominal p-value | 0.002352941 |
| FDR q-value | 0.21529627 |
| FWER p-Value | 0.366 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NFKB2 | 23810 | 65 | 2.638 | 0.0510 | Yes | ||
| 2 | MAP3K7IP2 | 19827 | 222 | 2.117 | 0.0863 | Yes | ||
| 3 | PIK3CG | 6635 | 265 | 2.027 | 0.1260 | Yes | ||
| 4 | CD86 | 1660 22601 | 300 | 1.921 | 0.1638 | Yes | ||
| 5 | IRAK1 | 4916 | 322 | 1.872 | 0.2014 | Yes | ||
| 6 | MAPK14 | 23313 | 334 | 1.857 | 0.2392 | Yes | ||
| 7 | TIRAP | 3075 19187 | 378 | 1.797 | 0.2740 | Yes | ||
| 8 | TBK1 | 12137 | 459 | 1.679 | 0.3044 | Yes | ||
| 9 | IKBKB | 4907 | 558 | 1.550 | 0.3311 | Yes | ||
| 10 | TRAF3 | 21147 | 648 | 1.428 | 0.3558 | Yes | ||
| 11 | IRF5 | 17502 | 711 | 1.348 | 0.3803 | Yes | ||
| 12 | MAP2K1 | 19082 | 767 | 1.285 | 0.4039 | Yes | ||
| 13 | IRF3 | 2149 2188 18255 | 891 | 1.169 | 0.4214 | Yes | ||
| 14 | MAPK3 | 6458 11170 | 961 | 1.090 | 0.4402 | Yes | ||
| 15 | MAP3K7 | 16255 | 1018 | 1.035 | 0.4586 | Yes | ||
| 16 | MAP3K7IP1 | 2193 2171 22419 | 1041 | 1.019 | 0.4785 | Yes | ||
| 17 | NFKBIA | 21065 | 1178 | 0.947 | 0.4907 | Yes | ||
| 18 | TLR7 | 24004 | 1274 | 0.878 | 0.5037 | Yes | ||
| 19 | RELA | 23783 | 1277 | 0.876 | 0.5217 | Yes | ||
| 20 | MAP3K8 | 23495 | 1281 | 0.872 | 0.5396 | Yes | ||
| 21 | CHUK | 23665 | 1310 | 0.853 | 0.5557 | Yes | ||
| 22 | IL1B | 14431 | 1311 | 0.852 | 0.5733 | Yes | ||
| 23 | LY96 | 14292 | 1440 | 0.773 | 0.5824 | Yes | ||
| 24 | IL12A | 4913 | 1462 | 0.763 | 0.5970 | Yes | ||
| 25 | MAP2K2 | 19933 | 1669 | 0.648 | 0.5993 | Yes | ||
| 26 | MAP2K3 | 20856 | 1692 | 0.634 | 0.6112 | Yes | ||
| 27 | CASP8 | 8694 | 1829 | 0.549 | 0.6152 | Yes | ||
| 28 | CCL5 | 20320 | 2011 | 0.454 | 0.6148 | No | ||
| 29 | FOS | 21202 | 2168 | 0.392 | 0.6145 | No | ||
| 30 | MAPK12 | 2182 2215 22164 | 2721 | 0.246 | 0.5898 | No | ||
| 31 | IFNA14 | 11917 | 2819 | 0.230 | 0.5893 | No | ||
| 32 | PIK3CD | 9563 | 2973 | 0.204 | 0.5852 | No | ||
| 33 | RIPK1 | 5381 | 3213 | 0.170 | 0.5758 | No | ||
| 34 | IKBKG | 2570 2562 4908 | 3736 | 0.119 | 0.5501 | No | ||
| 35 | TLR1 | 10190 | 3907 | 0.108 | 0.5432 | No | ||
| 36 | CD14 | 23452 | 3922 | 0.107 | 0.5446 | No | ||
| 37 | SPP1 | 5501 | 3949 | 0.105 | 0.5454 | No | ||
| 38 | CCL4 | 1362 20730 | 4072 | 0.097 | 0.5408 | No | ||
| 39 | MAPK9 | 1233 20903 1383 | 4094 | 0.096 | 0.5416 | No | ||
| 40 | MAPK13 | 23314 | 4397 | 0.080 | 0.5270 | No | ||
| 41 | LBP | 14760 | 4581 | 0.073 | 0.5186 | No | ||
| 42 | IRF7 | 12008 | 4613 | 0.072 | 0.5184 | No | ||
| 43 | IL6 | 16895 | 4753 | 0.067 | 0.5123 | No | ||
| 44 | IFNA4 | 9150 | 4930 | 0.061 | 0.5041 | No | ||
| 45 | TLR8 | 9308 | 5139 | 0.055 | 0.4940 | No | ||
| 46 | CCL3 | 20317 | 5509 | 0.046 | 0.4750 | No | ||
| 47 | TLR9 | 19331 3127 | 5777 | 0.040 | 0.4614 | No | ||
| 48 | IFNAR1 | 22703 | 5916 | 0.037 | 0.4547 | No | ||
| 49 | PIK3R5 | 11507 | 5998 | 0.036 | 0.4511 | No | ||
| 50 | MAPK11 | 2264 9618 22163 | 6170 | 0.033 | 0.4425 | No | ||
| 51 | TNF | 23004 | 6399 | 0.029 | 0.4308 | No | ||
| 52 | CXCL11 | 16478 | 7227 | 0.017 | 0.3865 | No | ||
| 53 | IFNA7 | 9152 | 7241 | 0.017 | 0.3862 | No | ||
| 54 | IL12B | 20918 | 7350 | 0.016 | 0.3807 | No | ||
| 55 | PIK3R1 | 3170 | 7780 | 0.010 | 0.3577 | No | ||
| 56 | IFNB1 | 15846 | 8029 | 0.007 | 0.3445 | No | ||
| 57 | TLR5 | 14017 | 8983 | -0.004 | 0.2931 | No | ||
| 58 | IFNA13 | 15844 | 9414 | -0.009 | 0.2701 | No | ||
| 59 | AKT1 | 8568 | 9662 | -0.013 | 0.2570 | No | ||
| 60 | TICAM2 | 23440 | 9962 | -0.016 | 0.2412 | No | ||
| 61 | AKT2 | 4365 4366 | 11247 | -0.035 | 0.1726 | No | ||
| 62 | IRAK4 | 22379 11185 | 11420 | -0.038 | 0.1641 | No | ||
| 63 | PIK3CA | 9562 | 11493 | -0.039 | 0.1610 | No | ||
| 64 | MAP2K7 | 6453 | 11507 | -0.039 | 0.1611 | No | ||
| 65 | IFNA2 | 9149 | 12208 | -0.054 | 0.1244 | No | ||
| 66 | CXCL10 | 16477 | 12263 | -0.055 | 0.1227 | No | ||
| 67 | TOLLIP | 7039 12038 | 12281 | -0.056 | 0.1229 | No | ||
| 68 | TLR6 | 215 16528 | 12343 | -0.057 | 0.1208 | No | ||
| 69 | IFNAR2 | 22705 1699 | 12364 | -0.058 | 0.1209 | No | ||
| 70 | PIK3R2 | 18850 | 12662 | -0.067 | 0.1062 | No | ||
| 71 | TLR3 | 18884 | 13002 | -0.078 | 0.0896 | No | ||
| 72 | IFNA6 | 17749 | 13130 | -0.083 | 0.0844 | No | ||
| 73 | CXCL9 | 5099 | 13278 | -0.089 | 0.0783 | No | ||
| 74 | IFNA1 | 9147 | 13306 | -0.090 | 0.0787 | No | ||
| 75 | MAPK10 | 11169 | 13511 | -0.099 | 0.0697 | No | ||
| 76 | PIK3R3 | 5248 | 13606 | -0.105 | 0.0668 | No | ||
| 77 | IFNA5 | 9151 | 13628 | -0.106 | 0.0679 | No | ||
| 78 | NFKB1 | 15160 | 13771 | -0.113 | 0.0625 | No | ||
| 79 | CD40 | 10207 2961 2968 5783 | 14056 | -0.131 | 0.0499 | No | ||
| 80 | TLR4 | 2329 10191 5770 | 14171 | -0.139 | 0.0466 | No | ||
| 81 | IKBKE | 67 | 14176 | -0.139 | 0.0493 | No | ||
| 82 | MAP2K6 | 20614 1414 | 14512 | -0.170 | 0.0347 | No | ||
| 83 | FADD | 17536 8950 4711 | 14843 | -0.208 | 0.0212 | No | ||
| 84 | MYD88 | 18970 | 15953 | -0.417 | -0.0300 | No | ||
| 85 | STAT1 | 3936 5524 | 15957 | -0.419 | -0.0215 | No | ||
| 86 | CD80 | 22758 | 16016 | -0.435 | -0.0157 | No | ||
| 87 | RAC1 | 16302 | 16377 | -0.547 | -0.0238 | No | ||
| 88 | TLR2 | 15308 | 16556 | -0.614 | -0.0208 | No | ||
| 89 | MAPK8 | 6459 | 16644 | -0.646 | -0.0121 | No | ||
| 90 | JUN | 15832 | 16667 | -0.656 | 0.0003 | No | ||
| 91 | MAP2K4 | 20405 | 16860 | -0.753 | 0.0055 | No | ||
| 92 | PIK3CB | 19030 | 17167 | -0.904 | 0.0076 | No | ||
| 93 | TRAF6 | 5797 14940 | 17525 | -1.076 | 0.0106 | No | ||
| 94 | MAPK1 | 1642 11167 | 17667 | -1.160 | 0.0270 | No | ||
| 95 | AKT3 | 13739 982 | 17683 | -1.172 | 0.0504 | No |