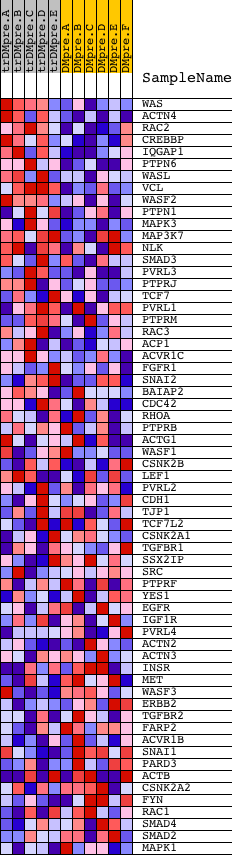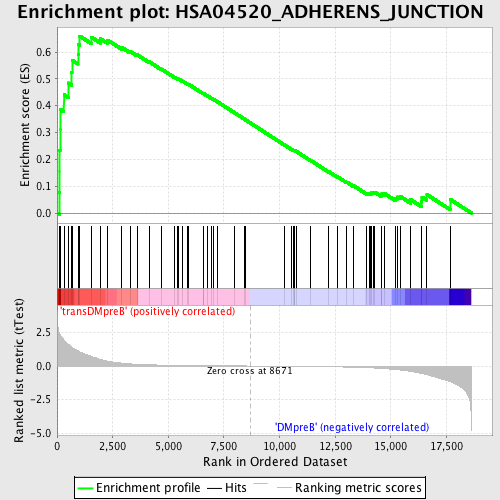
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | HSA04520_ADHERENS_JUNCTION |
| Enrichment Score (ES) | 0.6593918 |
| Normalized Enrichment Score (NES) | 1.711347 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.27055365 |
| FWER p-Value | 0.173 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | WAS | 24193 | 99 | 2.439 | 0.0759 | Yes | ||
| 2 | ACTN4 | 17905 1798 1983 | 116 | 2.388 | 0.1547 | Yes | ||
| 3 | RAC2 | 22217 | 117 | 2.387 | 0.2342 | Yes | ||
| 4 | CREBBP | 22682 8783 | 133 | 2.312 | 0.3104 | Yes | ||
| 5 | IQGAP1 | 6619 | 136 | 2.288 | 0.3866 | Yes | ||
| 6 | PTPN6 | 17002 | 309 | 1.905 | 0.4408 | Yes | ||
| 7 | WASL | 17203 | 501 | 1.629 | 0.4848 | Yes | ||
| 8 | VCL | 22083 | 654 | 1.422 | 0.5240 | Yes | ||
| 9 | WASF2 | 6326 | 671 | 1.401 | 0.5698 | Yes | ||
| 10 | PTPN1 | 5325 | 939 | 1.121 | 0.5927 | Yes | ||
| 11 | MAPK3 | 6458 11170 | 961 | 1.090 | 0.6279 | Yes | ||
| 12 | MAP3K7 | 16255 | 1018 | 1.035 | 0.6594 | Yes | ||
| 13 | NLK | 5179 5178 | 1529 | 0.723 | 0.6560 | No | ||
| 14 | SMAD3 | 19084 | 1948 | 0.484 | 0.6496 | No | ||
| 15 | PVRL3 | 1674 22579 1675 | 2262 | 0.365 | 0.6449 | No | ||
| 16 | PTPRJ | 9664 | 2913 | 0.213 | 0.6169 | No | ||
| 17 | TCF7 | 1467 20466 | 3291 | 0.161 | 0.6019 | No | ||
| 18 | PVRL1 | 3006 19482 12211 7192 | 3595 | 0.131 | 0.5900 | No | ||
| 19 | PTPRM | 22904 | 4132 | 0.094 | 0.5642 | No | ||
| 20 | RAC3 | 20561 | 4683 | 0.069 | 0.5369 | No | ||
| 21 | ACP1 | 4317 | 5276 | 0.051 | 0.5067 | No | ||
| 22 | ACVR1C | 14585 | 5425 | 0.048 | 0.5003 | No | ||
| 23 | FGFR1 | 3789 8968 | 5469 | 0.047 | 0.4995 | No | ||
| 24 | SNAI2 | 22848 | 5632 | 0.043 | 0.4922 | No | ||
| 25 | BAIAP2 | 449 4236 | 5879 | 0.038 | 0.4802 | No | ||
| 26 | CDC42 | 4503 8722 4504 2465 | 5895 | 0.038 | 0.4807 | No | ||
| 27 | RHOA | 8624 4409 4410 | 6560 | 0.027 | 0.4458 | No | ||
| 28 | PTPRB | 19875 | 6567 | 0.027 | 0.4463 | No | ||
| 29 | ACTG1 | 8535 | 6767 | 0.024 | 0.4364 | No | ||
| 30 | WASF1 | 13514 | 6956 | 0.021 | 0.4270 | No | ||
| 31 | CSNK2B | 23008 | 7009 | 0.020 | 0.4248 | No | ||
| 32 | LEF1 | 1860 15420 | 7190 | 0.018 | 0.4157 | No | ||
| 33 | PVRL2 | 9671 5336 | 7985 | 0.008 | 0.3732 | No | ||
| 34 | CDH1 | 18479 | 8400 | 0.003 | 0.3510 | No | ||
| 35 | TJP1 | 17803 | 8481 | 0.002 | 0.3467 | No | ||
| 36 | TCF7L2 | 10048 5646 | 10216 | -0.020 | 0.2539 | No | ||
| 37 | CSNK2A1 | 14797 | 10544 | -0.024 | 0.2371 | No | ||
| 38 | TGFBR1 | 16213 10165 5745 | 10621 | -0.025 | 0.2339 | No | ||
| 39 | SSX2IP | 13617 | 10657 | -0.026 | 0.2328 | No | ||
| 40 | SRC | 5507 | 10688 | -0.026 | 0.2321 | No | ||
| 41 | PTPRF | 5331 2545 | 10768 | -0.027 | 0.2288 | No | ||
| 42 | YES1 | 5930 | 11369 | -0.037 | 0.1976 | No | ||
| 43 | EGFR | 1329 20944 | 12205 | -0.054 | 0.1544 | No | ||
| 44 | IGF1R | 9157 | 12594 | -0.064 | 0.1357 | No | ||
| 45 | PVRL4 | 14055 970 | 13017 | -0.079 | 0.1156 | No | ||
| 46 | ACTN2 | 21545 | 13310 | -0.090 | 0.1028 | No | ||
| 47 | ACTN3 | 23967 8540 | 13926 | -0.122 | 0.0737 | No | ||
| 48 | INSR | 18950 | 14023 | -0.129 | 0.0729 | No | ||
| 49 | MET | 17520 | 14073 | -0.132 | 0.0746 | No | ||
| 50 | WASF3 | 16625 | 14144 | -0.137 | 0.0754 | No | ||
| 51 | ERBB2 | 8913 | 14207 | -0.142 | 0.0768 | No | ||
| 52 | TGFBR2 | 2994 3001 10167 | 14270 | -0.147 | 0.0784 | No | ||
| 53 | FARP2 | 14175 | 14565 | -0.176 | 0.0684 | No | ||
| 54 | ACVR1B | 4335 22353 | 14601 | -0.180 | 0.0725 | No | ||
| 55 | SNAI1 | 14726 | 14705 | -0.191 | 0.0733 | No | ||
| 56 | PARD3 | 8212 13553 | 15205 | -0.256 | 0.0549 | No | ||
| 57 | ACTB | 8534 337 337 338 | 15280 | -0.269 | 0.0599 | No | ||
| 58 | CSNK2A2 | 4567 8808 | 15415 | -0.291 | 0.0624 | No | ||
| 59 | FYN | 3375 3395 20052 | 15886 | -0.398 | 0.0503 | No | ||
| 60 | RAC1 | 16302 | 16377 | -0.547 | 0.0421 | No | ||
| 61 | SMAD4 | 5058 | 16399 | -0.553 | 0.0594 | No | ||
| 62 | SMAD2 | 23511 | 16624 | -0.640 | 0.0686 | No | ||
| 63 | MAPK1 | 1642 11167 | 17667 | -1.160 | 0.0511 | No |