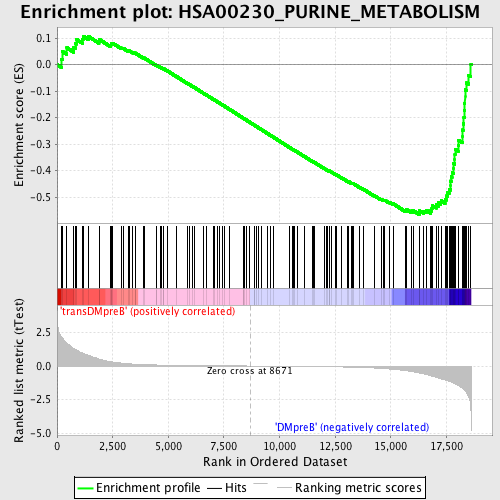
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | DMpreB |
| GeneSet | HSA00230_PURINE_METABOLISM |
| Enrichment Score (ES) | -0.5657734 |
| Normalized Enrichment Score (NES) | -1.548237 |
| Nominal p-value | 0.0017636684 |
| FDR q-value | 0.21280041 |
| FWER p-Value | 0.909 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | POLD3 | 17742 | 204 | 2.147 | 0.0205 | No | ||
| 2 | HPRT1 | 2655 24339 408 | 244 | 2.067 | 0.0487 | No | ||
| 3 | PKM2 | 3642 9573 | 439 | 1.709 | 0.0633 | No | ||
| 4 | POLR1A | 9749 5393 | 745 | 1.304 | 0.0659 | No | ||
| 5 | POLA2 | 23988 | 819 | 1.240 | 0.0802 | No | ||
| 6 | AK2 | 8563 2479 16073 | 873 | 1.189 | 0.0947 | No | ||
| 7 | POLE | 16755 | 1140 | 0.981 | 0.0947 | No | ||
| 8 | POLD1 | 17847 | 1169 | 0.959 | 0.1073 | No | ||
| 9 | POLD4 | 12822 | 1404 | 0.798 | 0.1063 | No | ||
| 10 | NT5C | 20151 | 1883 | 0.516 | 0.0881 | No | ||
| 11 | PRPS2 | 24003 | 1912 | 0.501 | 0.0939 | No | ||
| 12 | PDE8A | 18202 | 2417 | 0.317 | 0.0713 | No | ||
| 13 | XDH | 22897 | 2423 | 0.315 | 0.0756 | No | ||
| 14 | PDE2A | 18170 | 2430 | 0.312 | 0.0799 | No | ||
| 15 | RRM1 | 18163 | 2497 | 0.295 | 0.0806 | No | ||
| 16 | RRM2 | 5401 5400 | 2892 | 0.217 | 0.0625 | No | ||
| 17 | NME1 | 9467 | 2961 | 0.206 | 0.0619 | No | ||
| 18 | PDE7A | 9544 1788 | 3212 | 0.170 | 0.0508 | No | ||
| 19 | ECGF1 | 22160 | 3245 | 0.166 | 0.0515 | No | ||
| 20 | CANT1 | 13304 | 3395 | 0.148 | 0.0457 | No | ||
| 21 | PDE4D | 10722 6235 | 3504 | 0.138 | 0.0418 | No | ||
| 22 | POLR3A | 21900 | 3508 | 0.137 | 0.0437 | No | ||
| 23 | NT5E | 19360 18702 | 3904 | 0.108 | 0.0239 | No | ||
| 24 | AMPD1 | 5051 | 3916 | 0.107 | 0.0249 | No | ||
| 25 | POLR2C | 9750 | 4458 | 0.078 | -0.0032 | No | ||
| 26 | ENTPD6 | 4496 2678 | 4460 | 0.077 | -0.0022 | No | ||
| 27 | GDA | 4764 | 4626 | 0.071 | -0.0100 | No | ||
| 28 | PDE6H | 17257 | 4689 | 0.069 | -0.0124 | No | ||
| 29 | ITPA | 9193 | 4759 | 0.066 | -0.0151 | No | ||
| 30 | ADCY8 | 22281 | 4797 | 0.065 | -0.0162 | No | ||
| 31 | NPR2 | 16230 | 4955 | 0.060 | -0.0238 | No | ||
| 32 | PAPSS2 | 6258 | 5386 | 0.049 | -0.0463 | No | ||
| 33 | POLE2 | 21053 | 5882 | 0.038 | -0.0726 | No | ||
| 34 | ADCY4 | 2980 21818 | 5954 | 0.036 | -0.0759 | No | ||
| 35 | PDE4C | 8481 | 6098 | 0.034 | -0.0831 | No | ||
| 36 | ADCY9 | 22680 | 6166 | 0.033 | -0.0862 | No | ||
| 37 | ENTPD2 | 8713 | 6584 | 0.026 | -0.1084 | No | ||
| 38 | AMPD2 | 1931 15197 | 6692 | 0.025 | -0.1138 | No | ||
| 39 | PKLR | 1850 15545 | 7037 | 0.020 | -0.1321 | No | ||
| 40 | POLR3K | 12447 7372 | 7063 | 0.019 | -0.1332 | No | ||
| 41 | IMPDH1 | 17197 1131 | 7187 | 0.018 | -0.1396 | No | ||
| 42 | NT5C1B | 21315 | 7311 | 0.016 | -0.1460 | No | ||
| 43 | POLE3 | 7200 | 7444 | 0.014 | -0.1529 | No | ||
| 44 | GUCY1A3 | 7216 12245 15310 | 7505 | 0.013 | -0.1560 | No | ||
| 45 | GUCY2F | 24045 | 7762 | 0.010 | -0.1697 | No | ||
| 46 | ENTPD3 | 19266 | 8358 | 0.004 | -0.2018 | No | ||
| 47 | ENTPD8 | 12996 | 8370 | 0.004 | -0.2024 | No | ||
| 48 | PDE10A | 23387 | 8440 | 0.003 | -0.2061 | No | ||
| 49 | NPR1 | 9480 | 8528 | 0.002 | -0.2107 | No | ||
| 50 | AK5 | 6042 | 8635 | 0.000 | -0.2165 | No | ||
| 51 | ALLC | 21093 2130 | 8861 | -0.002 | -0.2286 | No | ||
| 52 | GUCY1A2 | 19580 | 8869 | -0.002 | -0.2289 | No | ||
| 53 | POLA1 | 24112 | 8962 | -0.003 | -0.2339 | No | ||
| 54 | NT5M | 8345 4175 | 9073 | -0.005 | -0.2398 | No | ||
| 55 | PDE1C | 5235 | 9189 | -0.006 | -0.2459 | No | ||
| 56 | GUCY2C | 1140 16954 | 9454 | -0.010 | -0.2600 | No | ||
| 57 | POLR2B | 16817 | 9610 | -0.012 | -0.2682 | No | ||
| 58 | ENPP3 | 19803 | 9728 | -0.013 | -0.2744 | No | ||
| 59 | RRM2B | 6902 6903 11801 | 10432 | -0.023 | -0.3121 | No | ||
| 60 | ADCY5 | 10312 | 10596 | -0.025 | -0.3205 | No | ||
| 61 | PDE7B | 19807 | 10626 | -0.026 | -0.3217 | No | ||
| 62 | PDE11A | 10805 | 10649 | -0.026 | -0.3225 | No | ||
| 63 | PDE1A | 9541 5234 | 10813 | -0.028 | -0.3309 | No | ||
| 64 | ADK | 3302 3454 8555 | 11121 | -0.033 | -0.3470 | No | ||
| 65 | ADCY6 | 22139 2283 8551 | 11490 | -0.039 | -0.3664 | No | ||
| 66 | ENTPD1 | 4495 | 11525 | -0.039 | -0.3676 | No | ||
| 67 | POLR3H | 13460 8135 | 11584 | -0.041 | -0.3702 | No | ||
| 68 | PDE4A | 3037 19542 3083 3023 | 11997 | -0.049 | -0.3917 | No | ||
| 69 | POLR2A | 5394 | 12113 | -0.052 | -0.3972 | No | ||
| 70 | PRIM1 | 19847 | 12164 | -0.053 | -0.3991 | No | ||
| 71 | DGUOK | 17099 1041 | 12229 | -0.055 | -0.4018 | No | ||
| 72 | PDE9A | 23301 | 12232 | -0.055 | -0.4011 | No | ||
| 73 | GMPR | 21651 | 12338 | -0.057 | -0.4059 | No | ||
| 74 | AK1 | 4363 | 12500 | -0.061 | -0.4137 | No | ||
| 75 | PDE6G | 20119 | 12577 | -0.064 | -0.4169 | No | ||
| 76 | PDE5A | 15431 | 12802 | -0.071 | -0.4280 | No | ||
| 77 | AMPD3 | 4383 2110 | 13060 | -0.080 | -0.4407 | No | ||
| 78 | ADCY7 | 8552 448 | 13118 | -0.082 | -0.4426 | No | ||
| 79 | PDE8B | 5756 | 13249 | -0.087 | -0.4483 | No | ||
| 80 | GUCY1B3 | 15311 | 13256 | -0.087 | -0.4474 | No | ||
| 81 | NUDT9 | 7897 | 13302 | -0.090 | -0.4485 | No | ||
| 82 | ADCY2 | 21412 | 13602 | -0.104 | -0.4631 | No | ||
| 83 | PDE3B | 5236 | 13786 | -0.114 | -0.4714 | No | ||
| 84 | IMPDH2 | 10730 | 14256 | -0.146 | -0.4946 | No | ||
| 85 | GMPR2 | 2711 | 14558 | -0.175 | -0.5083 | No | ||
| 86 | NME7 | 926 4101 14070 3986 3946 931 | 14664 | -0.187 | -0.5112 | No | ||
| 87 | AK3L1 | 8564 | 14724 | -0.193 | -0.5116 | No | ||
| 88 | PNPT1 | 20936 | 14928 | -0.219 | -0.5194 | No | ||
| 89 | ADCY3 | 21329 894 | 15106 | -0.241 | -0.5254 | No | ||
| 90 | PAICS | 16820 | 15673 | -0.349 | -0.5509 | No | ||
| 91 | RFC5 | 13005 7791 | 15700 | -0.356 | -0.5471 | No | ||
| 92 | PDE4B | 9543 | 15910 | -0.406 | -0.5524 | No | ||
| 93 | POLE4 | 12441 | 16004 | -0.432 | -0.5511 | No | ||
| 94 | POLR2E | 3325 19699 | 16276 | -0.508 | -0.5583 | Yes | ||
| 95 | GART | 22543 1754 | 16281 | -0.510 | -0.5511 | Yes | ||
| 96 | PPAT | 6081 | 16471 | -0.577 | -0.5528 | Yes | ||
| 97 | ENPP1 | 19804 | 16584 | -0.626 | -0.5497 | Yes | ||
| 98 | PDE6D | 13897 3945 | 16796 | -0.717 | -0.5506 | Yes | ||
| 99 | ENTPD5 | 4497 | 16843 | -0.741 | -0.5422 | Yes | ||
| 100 | ENTPD4 | 7449 | 16858 | -0.750 | -0.5320 | Yes | ||
| 101 | PAPSS1 | 15419 | 17035 | -0.850 | -0.5290 | Yes | ||
| 102 | POLR2J | 16672 | 17132 | -0.889 | -0.5211 | Yes | ||
| 103 | ADSL | 4358 | 17255 | -0.952 | -0.5138 | Yes | ||
| 104 | ATIC | 14231 3968 | 17439 | -1.030 | -0.5086 | Yes | ||
| 105 | ADSSL1 | 21136 2087 | 17492 | -1.059 | -0.4958 | Yes | ||
| 106 | ZNRD1 | 1491 22987 | 17540 | -1.081 | -0.4825 | Yes | ||
| 107 | POLR3B | 12875 | 17620 | -1.135 | -0.4701 | Yes | ||
| 108 | POLD2 | 20537 | 17687 | -1.174 | -0.4565 | Yes | ||
| 109 | POLR2I | 12839 | 17691 | -1.175 | -0.4394 | Yes | ||
| 110 | POLR1B | 14857 | 17713 | -1.191 | -0.4231 | Yes | ||
| 111 | NT5C2 | 3768 8052 | 17752 | -1.221 | -0.4072 | Yes | ||
| 112 | NUDT2 | 16239 | 17821 | -1.266 | -0.3923 | Yes | ||
| 113 | PRPS1 | 24233 | 17827 | -1.272 | -0.3739 | Yes | ||
| 114 | ADA | 2703 14361 | 17876 | -1.307 | -0.3573 | Yes | ||
| 115 | DCK | 16808 | 17881 | -1.308 | -0.3384 | Yes | ||
| 116 | FHIT | 21920 | 17890 | -1.314 | -0.3195 | Yes | ||
| 117 | GMPS | 15578 | 18028 | -1.431 | -0.3060 | Yes | ||
| 118 | GUK1 | 20432 | 18041 | -1.442 | -0.2854 | Yes | ||
| 119 | APRT | 8620 | 18225 | -1.660 | -0.2710 | Yes | ||
| 120 | NME6 | 3139 19297 | 18231 | -1.669 | -0.2468 | Yes | ||
| 121 | NT5C3 | 17136 | 18251 | -1.698 | -0.2229 | Yes | ||
| 122 | POLR1D | 3593 3658 16623 | 18284 | -1.749 | -0.1990 | Yes | ||
| 123 | NME2 | 9468 | 18309 | -1.791 | -0.1740 | Yes | ||
| 124 | POLR2H | 10888 | 18321 | -1.818 | -0.1479 | Yes | ||
| 125 | NME4 | 23065 1577 | 18334 | -1.844 | -0.1215 | Yes | ||
| 126 | NUDT5 | 15121 | 18336 | -1.845 | -0.0945 | Yes | ||
| 127 | POLR2G | 23753 | 18407 | -1.978 | -0.0693 | Yes | ||
| 128 | NP | 22027 9597 5273 5274 | 18493 | -2.234 | -0.0411 | Yes | ||
| 129 | POLR2K | 9413 | 18599 | -3.250 | 0.0009 | Yes |

