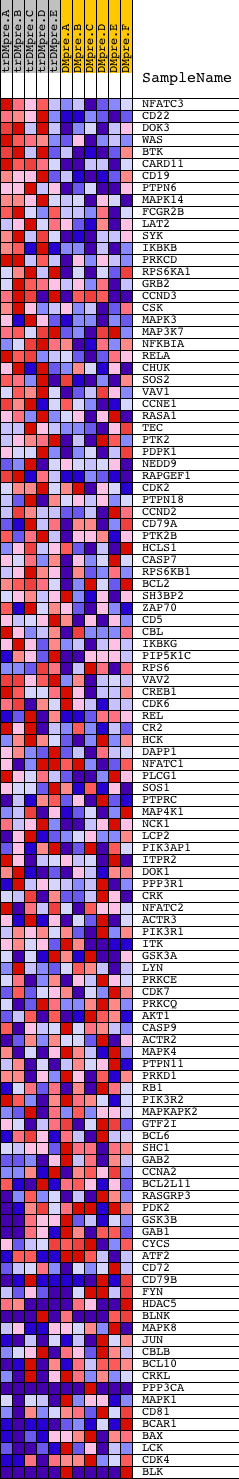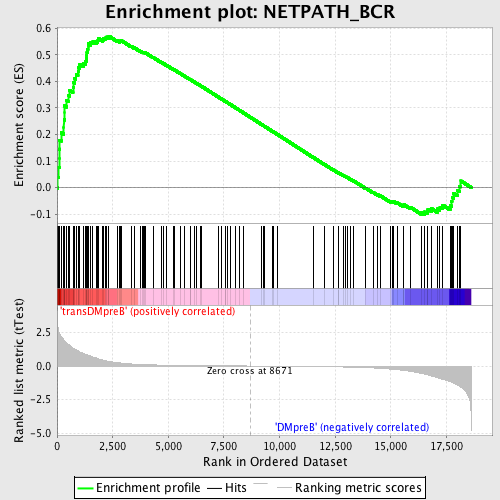
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | NETPATH_BCR |
| Enrichment Score (ES) | 0.57013327 |
| Normalized Enrichment Score (NES) | 1.6015425 |
| Nominal p-value | 0.002247191 |
| FDR q-value | 0.23095824 |
| FWER p-Value | 0.892 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NFATC3 | 5169 | 38 | 2.872 | 0.0395 | Yes | ||
| 2 | CD22 | 17881 | 70 | 2.621 | 0.0757 | Yes | ||
| 3 | DOK3 | 3229 21451 | 95 | 2.483 | 0.1103 | Yes | ||
| 4 | WAS | 24193 | 99 | 2.439 | 0.1454 | Yes | ||
| 5 | BTK | 24061 | 118 | 2.378 | 0.1788 | Yes | ||
| 6 | CARD11 | 8436 | 186 | 2.186 | 0.2068 | Yes | ||
| 7 | CD19 | 17640 | 306 | 1.909 | 0.2280 | Yes | ||
| 8 | PTPN6 | 17002 | 309 | 1.905 | 0.2554 | Yes | ||
| 9 | MAPK14 | 23313 | 334 | 1.857 | 0.2810 | Yes | ||
| 10 | FCGR2B | 8959 | 336 | 1.855 | 0.3078 | Yes | ||
| 11 | LAT2 | 16351 | 424 | 1.728 | 0.3281 | Yes | ||
| 12 | SYK | 21636 | 513 | 1.619 | 0.3467 | Yes | ||
| 13 | IKBKB | 4907 | 558 | 1.550 | 0.3668 | Yes | ||
| 14 | PRKCD | 21897 | 716 | 1.338 | 0.3776 | Yes | ||
| 15 | RPS6KA1 | 15725 | 736 | 1.314 | 0.3956 | Yes | ||
| 16 | GRB2 | 20149 | 792 | 1.266 | 0.4109 | Yes | ||
| 17 | CCND3 | 4489 4490 | 849 | 1.205 | 0.4253 | Yes | ||
| 18 | CSK | 8805 | 940 | 1.117 | 0.4366 | Yes | ||
| 19 | MAPK3 | 6458 11170 | 961 | 1.090 | 0.4513 | Yes | ||
| 20 | MAP3K7 | 16255 | 1018 | 1.035 | 0.4632 | Yes | ||
| 21 | NFKBIA | 21065 | 1178 | 0.947 | 0.4683 | Yes | ||
| 22 | RELA | 23783 | 1277 | 0.876 | 0.4757 | Yes | ||
| 23 | CHUK | 23665 | 1310 | 0.853 | 0.4863 | Yes | ||
| 24 | SOS2 | 21049 | 1331 | 0.845 | 0.4974 | Yes | ||
| 25 | VAV1 | 23173 | 1340 | 0.841 | 0.5092 | Yes | ||
| 26 | CCNE1 | 17857 | 1369 | 0.822 | 0.5195 | Yes | ||
| 27 | RASA1 | 10174 | 1390 | 0.808 | 0.5301 | Yes | ||
| 28 | TEC | 16514 | 1394 | 0.805 | 0.5416 | Yes | ||
| 29 | PTK2 | 22271 | 1498 | 0.741 | 0.5468 | Yes | ||
| 30 | PDPK1 | 23097 | 1610 | 0.675 | 0.5505 | Yes | ||
| 31 | NEDD9 | 3206 21483 | 1777 | 0.580 | 0.5500 | Yes | ||
| 32 | RAPGEF1 | 4218 2860 | 1832 | 0.544 | 0.5549 | Yes | ||
| 33 | CDK2 | 3438 3373 19592 3322 | 1865 | 0.529 | 0.5608 | Yes | ||
| 34 | PTPN18 | 14279 | 2046 | 0.440 | 0.5575 | Yes | ||
| 35 | CCND2 | 16987 | 2088 | 0.425 | 0.5614 | Yes | ||
| 36 | CD79A | 18342 | 2161 | 0.395 | 0.5632 | Yes | ||
| 37 | PTK2B | 21776 | 2197 | 0.385 | 0.5669 | Yes | ||
| 38 | HCLS1 | 22770 | 2314 | 0.350 | 0.5657 | Yes | ||
| 39 | CASP7 | 23826 | 2325 | 0.347 | 0.5701 | Yes | ||
| 40 | RPS6KB1 | 7815 1207 13040 | 2699 | 0.250 | 0.5536 | No | ||
| 41 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 2820 | 0.230 | 0.5504 | No | ||
| 42 | SH3BP2 | 16874 | 2850 | 0.225 | 0.5521 | No | ||
| 43 | ZAP70 | 14271 4042 | 2886 | 0.219 | 0.5534 | No | ||
| 44 | CD5 | 23741 | 3322 | 0.156 | 0.5322 | No | ||
| 45 | CBL | 19154 | 3468 | 0.140 | 0.5263 | No | ||
| 46 | IKBKG | 2570 2562 4908 | 3736 | 0.119 | 0.5136 | No | ||
| 47 | PIP5K1C | 5250 19929 | 3843 | 0.112 | 0.5095 | No | ||
| 48 | RPS6 | 9757 | 3898 | 0.108 | 0.5082 | No | ||
| 49 | VAV2 | 5848 2670 | 3906 | 0.108 | 0.5093 | No | ||
| 50 | CREB1 | 3990 8782 4558 4093 | 3961 | 0.105 | 0.5079 | No | ||
| 51 | CDK6 | 16600 | 4318 | 0.083 | 0.4899 | No | ||
| 52 | REL | 9716 | 4702 | 0.068 | 0.4702 | No | ||
| 53 | CR2 | 3942 13707 | 4795 | 0.065 | 0.4662 | No | ||
| 54 | HCK | 14787 | 4919 | 0.061 | 0.4604 | No | ||
| 55 | DAPP1 | 11159 6445 6446 | 5222 | 0.053 | 0.4448 | No | ||
| 56 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 5266 | 0.052 | 0.4433 | No | ||
| 57 | PLCG1 | 14753 | 5539 | 0.045 | 0.4292 | No | ||
| 58 | SOS1 | 5476 | 5727 | 0.041 | 0.4197 | No | ||
| 59 | PTPRC | 5327 9662 | 5973 | 0.036 | 0.4070 | No | ||
| 60 | MAP4K1 | 18313 | 6010 | 0.036 | 0.4055 | No | ||
| 61 | NCK1 | 9447 5152 | 6189 | 0.033 | 0.3964 | No | ||
| 62 | LCP2 | 4988 9268 | 6264 | 0.031 | 0.3929 | No | ||
| 63 | PIK3AP1 | 23679 | 6445 | 0.029 | 0.3835 | No | ||
| 64 | ITPR2 | 9194 | 6505 | 0.028 | 0.3808 | No | ||
| 65 | DOK1 | 17104 1018 1177 | 7255 | 0.017 | 0.3405 | No | ||
| 66 | PPP3R1 | 9611 489 | 7403 | 0.015 | 0.3328 | No | ||
| 67 | CRK | 4559 1249 | 7549 | 0.013 | 0.3251 | No | ||
| 68 | NFATC2 | 5168 2866 | 7581 | 0.013 | 0.3236 | No | ||
| 69 | ACTR3 | 13166 | 7669 | 0.012 | 0.3191 | No | ||
| 70 | PIK3R1 | 3170 | 7780 | 0.010 | 0.3133 | No | ||
| 71 | ITK | 4934 | 8021 | 0.007 | 0.3004 | No | ||
| 72 | GSK3A | 405 | 8181 | 0.006 | 0.2919 | No | ||
| 73 | LYN | 16281 | 8372 | 0.004 | 0.2817 | No | ||
| 74 | PRKCE | 9575 | 9170 | -0.006 | 0.2387 | No | ||
| 75 | CDK7 | 21365 | 9267 | -0.007 | 0.2336 | No | ||
| 76 | PRKCQ | 2873 2831 | 9311 | -0.008 | 0.2314 | No | ||
| 77 | AKT1 | 8568 | 9662 | -0.013 | 0.2127 | No | ||
| 78 | CASP9 | 16001 2410 2458 | 9739 | -0.014 | 0.2088 | No | ||
| 79 | ACTR2 | 20523 | 9911 | -0.016 | 0.1998 | No | ||
| 80 | MAPK4 | 23409 | 11523 | -0.039 | 0.1133 | No | ||
| 81 | PTPN11 | 5326 16391 9660 | 12023 | -0.049 | 0.0870 | No | ||
| 82 | PRKD1 | 2074 21075 | 12420 | -0.059 | 0.0665 | No | ||
| 83 | RB1 | 21754 | 12653 | -0.066 | 0.0549 | No | ||
| 84 | PIK3R2 | 18850 | 12662 | -0.067 | 0.0554 | No | ||
| 85 | MAPKAPK2 | 13838 | 12872 | -0.073 | 0.0452 | No | ||
| 86 | GTF2I | 16353 5491 9863 3280 3184 | 12945 | -0.076 | 0.0424 | No | ||
| 87 | BCL6 | 22624 | 13041 | -0.079 | 0.0384 | No | ||
| 88 | SHC1 | 9813 9812 5430 | 13187 | -0.085 | 0.0318 | No | ||
| 89 | GAB2 | 1821 18184 2025 | 13330 | -0.091 | 0.0254 | No | ||
| 90 | CCNA2 | 15357 | 13871 | -0.120 | -0.0020 | No | ||
| 91 | BCL2L11 | 2790 14861 | 14199 | -0.141 | -0.0177 | No | ||
| 92 | RASGRP3 | 10767 | 14421 | -0.162 | -0.0273 | No | ||
| 93 | PDK2 | 20285 | 14532 | -0.172 | -0.0307 | No | ||
| 94 | GSK3B | 22761 | 14988 | -0.227 | -0.0520 | No | ||
| 95 | GAB1 | 18828 | 15058 | -0.235 | -0.0524 | No | ||
| 96 | CYCS | 8821 | 15141 | -0.247 | -0.0532 | No | ||
| 97 | ATF2 | 4418 2759 | 15291 | -0.272 | -0.0574 | No | ||
| 98 | CD72 | 8718 | 15559 | -0.322 | -0.0671 | No | ||
| 99 | CD79B | 20185 1309 | 15588 | -0.328 | -0.0639 | No | ||
| 100 | FYN | 3375 3395 20052 | 15886 | -0.398 | -0.0742 | No | ||
| 101 | HDAC5 | 1480 20205 | 16384 | -0.548 | -0.0931 | No | ||
| 102 | BLNK | 23681 3691 | 16514 | -0.592 | -0.0915 | No | ||
| 103 | MAPK8 | 6459 | 16644 | -0.646 | -0.0891 | No | ||
| 104 | JUN | 15832 | 16667 | -0.656 | -0.0808 | No | ||
| 105 | CBLB | 5531 22734 | 16829 | -0.732 | -0.0790 | No | ||
| 106 | BCL10 | 15397 | 17084 | -0.870 | -0.0801 | No | ||
| 107 | CRKL | 4560 | 17208 | -0.926 | -0.0734 | No | ||
| 108 | PPP3CA | 1863 5284 | 17331 | -0.993 | -0.0656 | No | ||
| 109 | MAPK1 | 1642 11167 | 17667 | -1.160 | -0.0669 | No | ||
| 110 | CD81 | 8719 | 17724 | -1.201 | -0.0526 | No | ||
| 111 | BCAR1 | 18741 | 17751 | -1.220 | -0.0363 | No | ||
| 112 | BAX | 17832 | 17815 | -1.264 | -0.0215 | No | ||
| 113 | LCK | 15746 | 17991 | -1.398 | -0.0107 | No | ||
| 114 | CDK4 | 3424 19859 | 18093 | -1.498 | 0.0055 | No | ||
| 115 | BLK | 3205 21791 | 18152 | -1.571 | 0.0251 | No |