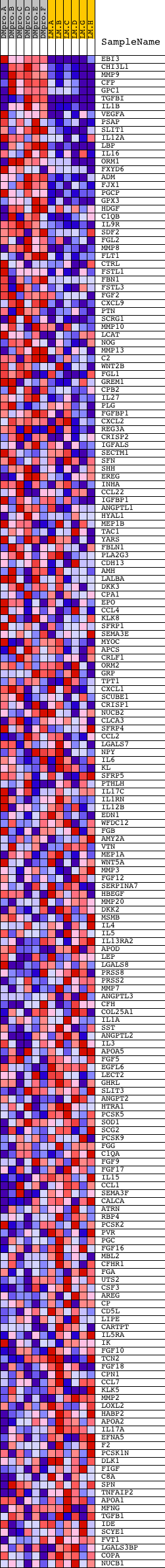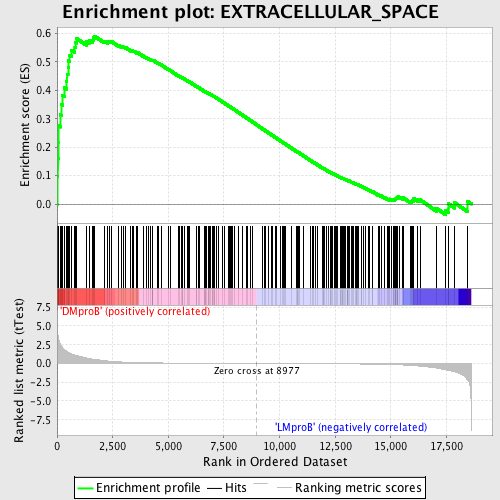
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | DMproB |
| GeneSet | EXTRACELLULAR_SPACE |
| Enrichment Score (ES) | 0.5909914 |
| Normalized Enrichment Score (NES) | 1.6235752 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.083983146 |
| FWER p-Value | 0.758 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EBI3 | 23193 | 4 | 5.900 | 0.0986 | Yes | ||
| 2 | CHI3L1 | 14131 | 39 | 3.745 | 0.1596 | Yes | ||
| 3 | MMP9 | 14732 | 54 | 3.526 | 0.2179 | Yes | ||
| 4 | CFP | 24174 | 58 | 3.500 | 0.2764 | Yes | ||
| 5 | GPC1 | 4796 | 158 | 2.551 | 0.3138 | Yes | ||
| 6 | TGFBI | 3181 21622 | 196 | 2.289 | 0.3501 | Yes | ||
| 7 | IL1B | 14431 | 253 | 2.069 | 0.3818 | Yes | ||
| 8 | VEGFA | 22969 | 322 | 1.878 | 0.4095 | Yes | ||
| 9 | PSAP | 20014 | 419 | 1.640 | 0.4318 | Yes | ||
| 10 | SLIT1 | 23678 | 444 | 1.596 | 0.4572 | Yes | ||
| 11 | IL12A | 4913 | 492 | 1.522 | 0.4802 | Yes | ||
| 12 | LBP | 14760 | 510 | 1.486 | 0.5042 | Yes | ||
| 13 | IL16 | 9171 | 572 | 1.376 | 0.5239 | Yes | ||
| 14 | ORM1 | 9516 | 640 | 1.273 | 0.5416 | Yes | ||
| 15 | FXYD6 | 19471 | 801 | 1.097 | 0.5513 | Yes | ||
| 16 | ADM | 18125 | 815 | 1.086 | 0.5688 | Yes | ||
| 17 | FJX1 | 4725 | 884 | 1.029 | 0.5824 | Yes | ||
| 18 | PGCP | 12030 | 1310 | 0.734 | 0.5716 | Yes | ||
| 19 | GPX3 | 20880 | 1437 | 0.666 | 0.5759 | Yes | ||
| 20 | HDGF | 9082 4846 4845 | 1595 | 0.583 | 0.5772 | Yes | ||
| 21 | C1QB | 8667 | 1623 | 0.569 | 0.5852 | Yes | ||
| 22 | IL9R | 20505 | 1685 | 0.541 | 0.5910 | Yes | ||
| 23 | SDF2 | 20762 | 2130 | 0.373 | 0.5731 | No | ||
| 24 | FGL2 | 16911 | 2273 | 0.333 | 0.5710 | No | ||
| 25 | MMP8 | 19570 | 2352 | 0.312 | 0.5720 | No | ||
| 26 | FLT1 | 3483 16287 | 2432 | 0.292 | 0.5726 | No | ||
| 27 | CTRL | 18760 | 2762 | 0.220 | 0.5584 | No | ||
| 28 | FSTL1 | 22765 | 2910 | 0.198 | 0.5538 | No | ||
| 29 | FBN1 | 14449 | 2987 | 0.189 | 0.5528 | No | ||
| 30 | FSTL3 | 19961 | 3077 | 0.176 | 0.5509 | No | ||
| 31 | FGF2 | 15608 | 3309 | 0.150 | 0.5409 | No | ||
| 32 | CXCL9 | 5099 | 3370 | 0.144 | 0.5401 | No | ||
| 33 | PTN | 5323 | 3443 | 0.137 | 0.5384 | No | ||
| 34 | SCRG1 | 18614 | 3577 | 0.124 | 0.5333 | No | ||
| 35 | MMP10 | 19571 | 3606 | 0.122 | 0.5338 | No | ||
| 36 | LCAT | 18762 | 3870 | 0.104 | 0.5213 | No | ||
| 37 | NOG | 13663 | 4035 | 0.094 | 0.5140 | No | ||
| 38 | MMP13 | 19573 3007 | 4122 | 0.090 | 0.5108 | No | ||
| 39 | C2 | 23013 | 4198 | 0.086 | 0.5082 | No | ||
| 40 | WNT2B | 15214 1838 | 4269 | 0.083 | 0.5058 | No | ||
| 41 | FGL1 | 18891 | 4290 | 0.082 | 0.5060 | No | ||
| 42 | GREM1 | 14485 | 4304 | 0.081 | 0.5067 | No | ||
| 43 | CPB2 | 21958 2894 3525 | 4524 | 0.073 | 0.4960 | No | ||
| 44 | IL27 | 17636 | 4541 | 0.072 | 0.4964 | No | ||
| 45 | PLG | 1593 23383 | 4673 | 0.068 | 0.4904 | No | ||
| 46 | FGFBP1 | 8967 | 4993 | 0.059 | 0.4741 | No | ||
| 47 | CXCL2 | 9790 | 5083 | 0.057 | 0.4702 | No | ||
| 48 | REG3A | 17406 | 5457 | 0.047 | 0.4507 | No | ||
| 49 | CRISP2 | 22979 | 5517 | 0.046 | 0.4483 | No | ||
| 50 | IGFALS | 9158 | 5605 | 0.045 | 0.4443 | No | ||
| 51 | SECTM1 | 20104 | 5618 | 0.045 | 0.4444 | No | ||
| 52 | SFN | 7060 12062 | 5653 | 0.044 | 0.4433 | No | ||
| 53 | SHH | 16581 | 5730 | 0.042 | 0.4399 | No | ||
| 54 | EREG | 4679 16797 | 5840 | 0.040 | 0.4346 | No | ||
| 55 | INHA | 14214 | 5893 | 0.039 | 0.4325 | No | ||
| 56 | CCL22 | 18518 | 5947 | 0.038 | 0.4302 | No | ||
| 57 | IGFBP1 | 20948 | 6258 | 0.033 | 0.4140 | No | ||
| 58 | ANGPTL1 | 10195 8589 13064 | 6259 | 0.033 | 0.4145 | No | ||
| 59 | HYAL1 | 9139 | 6352 | 0.031 | 0.4100 | No | ||
| 60 | MEP1B | 23612 5093 | 6381 | 0.031 | 0.4090 | No | ||
| 61 | TAC1 | 17526 19853 | 6608 | 0.027 | 0.3972 | No | ||
| 62 | YARS | 16071 | 6617 | 0.027 | 0.3972 | No | ||
| 63 | FBLN1 | 4716 2208 | 6666 | 0.026 | 0.3951 | No | ||
| 64 | PLA2G3 | 20968 | 6690 | 0.026 | 0.3943 | No | ||
| 65 | CDH13 | 4506 3826 | 6707 | 0.026 | 0.3938 | No | ||
| 66 | AMH | 19939 | 6783 | 0.025 | 0.3902 | No | ||
| 67 | LALBA | 22141 | 6802 | 0.025 | 0.3896 | No | ||
| 68 | DKK3 | 17673 2361 | 6867 | 0.024 | 0.3865 | No | ||
| 69 | CPA1 | 17496 | 6910 | 0.023 | 0.3847 | No | ||
| 70 | EPO | 8911 | 6977 | 0.022 | 0.3815 | No | ||
| 71 | CCL4 | 1362 20730 | 6983 | 0.022 | 0.3816 | No | ||
| 72 | KLK8 | 18269 18271 | 7020 | 0.022 | 0.3800 | No | ||
| 73 | SFRP1 | 9805 | 7057 | 0.021 | 0.3784 | No | ||
| 74 | SEMA3E | 16918 | 7060 | 0.021 | 0.3786 | No | ||
| 75 | MYOC | 9440 14079 | 7172 | 0.020 | 0.3729 | No | ||
| 76 | APCS | 13748 | 7261 | 0.019 | 0.3685 | No | ||
| 77 | CRLF1 | 18588 | 7419 | 0.017 | 0.3603 | No | ||
| 78 | ORM2 | 16193 | 7528 | 0.016 | 0.3547 | No | ||
| 79 | GRP | 23531 | 7713 | 0.014 | 0.3449 | No | ||
| 80 | TPT1 | 5799 | 7737 | 0.014 | 0.3439 | No | ||
| 81 | CXCL1 | 9044 | 7802 | 0.013 | 0.3406 | No | ||
| 82 | SCUBE1 | 7231 | 7827 | 0.012 | 0.3395 | No | ||
| 83 | CRISP1 | 22980 | 7904 | 0.012 | 0.3356 | No | ||
| 84 | NUCB2 | 18114 | 7981 | 0.011 | 0.3317 | No | ||
| 85 | CLCA3 | 6224 | 8150 | 0.009 | 0.3227 | No | ||
| 86 | SFRP4 | 9806 | 8313 | 0.007 | 0.3140 | No | ||
| 87 | CCL2 | 9788 | 8529 | 0.004 | 0.3024 | No | ||
| 88 | LGALS7 | 4992 | 8575 | 0.004 | 0.3000 | No | ||
| 89 | NPY | 17449 | 8670 | 0.003 | 0.2950 | No | ||
| 90 | IL6 | 16895 | 8787 | 0.002 | 0.2887 | No | ||
| 91 | KL | 16611 | 9220 | -0.003 | 0.2653 | No | ||
| 92 | SFRP5 | 23674 | 9303 | -0.004 | 0.2609 | No | ||
| 93 | PTHLH | 16931 | 9355 | -0.004 | 0.2582 | No | ||
| 94 | IL17C | 18438 | 9521 | -0.005 | 0.2494 | No | ||
| 95 | IL1RN | 15096 | 9652 | -0.007 | 0.2424 | No | ||
| 96 | IL12B | 20918 | 9683 | -0.007 | 0.2409 | No | ||
| 97 | EDN1 | 21658 | 9812 | -0.008 | 0.2341 | No | ||
| 98 | WFDC12 | 14356 | 9869 | -0.009 | 0.2312 | No | ||
| 99 | FGB | 15309 | 10046 | -0.010 | 0.2218 | No | ||
| 100 | AMY2A | 8587 | 10122 | -0.011 | 0.2179 | No | ||
| 101 | VTN | 20756 | 10171 | -0.012 | 0.2155 | No | ||
| 102 | MEP1A | 22974 | 10233 | -0.012 | 0.2124 | No | ||
| 103 | WNT5A | 22066 5880 | 10273 | -0.013 | 0.2105 | No | ||
| 104 | MMP3 | 19572 3148 | 10524 | -0.015 | 0.1972 | No | ||
| 105 | FGF12 | 1723 22621 | 10779 | -0.018 | 0.1837 | No | ||
| 106 | SERPINA7 | 6856 | 10788 | -0.019 | 0.1836 | No | ||
| 107 | HBEGF | 23457 | 10808 | -0.019 | 0.1829 | No | ||
| 108 | MMP20 | 19569 | 10838 | -0.019 | 0.1816 | No | ||
| 109 | DKK2 | 15418 | 10876 | -0.019 | 0.1799 | No | ||
| 110 | MSMB | 22053 | 11059 | -0.022 | 0.1704 | No | ||
| 111 | IL4 | 9174 | 11378 | -0.026 | 0.1536 | No | ||
| 112 | IL5 | 20884 10220 | 11462 | -0.027 | 0.1495 | No | ||
| 113 | IL13RA2 | 24036 | 11533 | -0.027 | 0.1462 | No | ||
| 114 | APOD | 22614 | 11632 | -0.029 | 0.1414 | No | ||
| 115 | LEP | 17505 | 11702 | -0.030 | 0.1381 | No | ||
| 116 | LGALS8 | 21544 | 11940 | -0.033 | 0.1258 | No | ||
| 117 | PRSS8 | 17606 | 11964 | -0.033 | 0.1251 | No | ||
| 118 | PRSS2 | 17474 | 11995 | -0.034 | 0.1241 | No | ||
| 119 | MMP7 | 19568 | 12030 | -0.034 | 0.1228 | No | ||
| 120 | ANGPTL3 | 16171 | 12119 | -0.036 | 0.1186 | No | ||
| 121 | CFH | 4515 13813 13814 | 12193 | -0.037 | 0.1153 | No | ||
| 122 | COL25A1 | 8056 1837 1844 1915 13376 | 12284 | -0.038 | 0.1110 | No | ||
| 123 | IL1A | 4915 | 12300 | -0.038 | 0.1109 | No | ||
| 124 | SST | 22625 | 12332 | -0.039 | 0.1098 | No | ||
| 125 | ANGPTL2 | 15032 18104 | 12377 | -0.040 | 0.1081 | No | ||
| 126 | IL3 | 20453 | 12445 | -0.041 | 0.1052 | No | ||
| 127 | APOA5 | 19469 | 12508 | -0.042 | 0.1025 | No | ||
| 128 | FGF5 | 16785 | 12536 | -0.042 | 0.1017 | No | ||
| 129 | EGFL6 | 24006 | 12610 | -0.044 | 0.0985 | No | ||
| 130 | LECT2 | 21443 | 12722 | -0.046 | 0.0932 | No | ||
| 131 | GHRL | 1183 17040 | 12748 | -0.046 | 0.0927 | No | ||
| 132 | SLIT3 | 20925 | 12777 | -0.046 | 0.0919 | No | ||
| 133 | ANGPT2 | 14885 3783 18923 | 12811 | -0.047 | 0.0909 | No | ||
| 134 | HTRA1 | 18051 | 12841 | -0.048 | 0.0901 | No | ||
| 135 | PCSK5 | 23714 | 12863 | -0.048 | 0.0898 | No | ||
| 136 | SOD1 | 9846 | 12930 | -0.049 | 0.0870 | No | ||
| 137 | SCG2 | 5410 14425 | 12960 | -0.050 | 0.0863 | No | ||
| 138 | PCSK9 | 15822 | 13043 | -0.052 | 0.0827 | No | ||
| 139 | FGG | 1891 | 13045 | -0.052 | 0.0835 | No | ||
| 140 | C1QA | 8666 | 13053 | -0.052 | 0.0840 | No | ||
| 141 | FGF9 | 8966 | 13117 | -0.053 | 0.0815 | No | ||
| 142 | FGF17 | 21757 | 13164 | -0.054 | 0.0799 | No | ||
| 143 | IL15 | 18826 3801 | 13242 | -0.056 | 0.0767 | No | ||
| 144 | CCL1 | 9787 | 13256 | -0.057 | 0.0769 | No | ||
| 145 | SEMA3F | 18999 3097 | 13307 | -0.058 | 0.0752 | No | ||
| 146 | CALCA | 4470 | 13398 | -0.061 | 0.0713 | No | ||
| 147 | ATRN | 4432 2964 | 13463 | -0.062 | 0.0689 | No | ||
| 148 | RBP4 | 23686 | 13468 | -0.063 | 0.0697 | No | ||
| 149 | PCSK2 | 9537 5231 14824 | 13494 | -0.063 | 0.0694 | No | ||
| 150 | PVR | 10020 | 13524 | -0.064 | 0.0689 | No | ||
| 151 | PGC | 23208 | 13673 | -0.069 | 0.0621 | No | ||
| 152 | FGF16 | 24270 | 13690 | -0.070 | 0.0624 | No | ||
| 153 | MBL2 | 23886 | 13780 | -0.073 | 0.0587 | No | ||
| 154 | CFHR1 | 11926 13815 | 13883 | -0.077 | 0.0545 | No | ||
| 155 | FGA | 1780 15566 | 14018 | -0.082 | 0.0486 | No | ||
| 156 | UTS2 | 15985 2520 | 14035 | -0.083 | 0.0491 | No | ||
| 157 | CSF3 | 1394 20671 | 14176 | -0.088 | 0.0430 | No | ||
| 158 | AREG | 16796 | 14185 | -0.088 | 0.0441 | No | ||
| 159 | CP | 4555 | 14423 | -0.099 | 0.0329 | No | ||
| 160 | CD5L | 15560 | 14504 | -0.104 | 0.0303 | No | ||
| 161 | LIPE | 17927 | 14563 | -0.107 | 0.0289 | No | ||
| 162 | CARTPT | 21374 | 14725 | -0.116 | 0.0221 | No | ||
| 163 | IL5RA | 17051 | 14865 | -0.125 | 0.0166 | No | ||
| 164 | IK | 23594 | 14915 | -0.129 | 0.0162 | No | ||
| 165 | FGF10 | 4719 8962 | 14928 | -0.130 | 0.0177 | No | ||
| 166 | TCN2 | 20550 | 15040 | -0.139 | 0.0140 | No | ||
| 167 | FGF18 | 4721 1351 | 15097 | -0.143 | 0.0133 | No | ||
| 168 | CPN1 | 23667 | 15109 | -0.145 | 0.0152 | No | ||
| 169 | CCL7 | 20743 | 15170 | -0.151 | 0.0144 | No | ||
| 170 | KLK5 | 1307 9237 | 15187 | -0.152 | 0.0161 | No | ||
| 171 | MMP2 | 18525 | 15210 | -0.154 | 0.0175 | No | ||
| 172 | LOXL2 | 8256 | 15228 | -0.156 | 0.0192 | No | ||
| 173 | HABP2 | 10364 23825 | 15239 | -0.157 | 0.0213 | No | ||
| 174 | APOA2 | 8615 4044 | 15285 | -0.162 | 0.0216 | No | ||
| 175 | IL17A | 14289 | 15316 | -0.165 | 0.0227 | No | ||
| 176 | EFNA5 | 4655 8884 | 15321 | -0.166 | 0.0253 | No | ||
| 177 | F2 | 14524 | 15389 | -0.174 | 0.0245 | No | ||
| 178 | PCSK1N | 6624 | 15541 | -0.195 | 0.0196 | No | ||
| 179 | DLK1 | 4632 | 15548 | -0.196 | 0.0226 | No | ||
| 180 | FIGF | 24212 | 15863 | -0.245 | 0.0096 | No | ||
| 181 | C8A | 15823 | 15941 | -0.259 | 0.0098 | No | ||
| 182 | SPN | 452 5493 | 15968 | -0.264 | 0.0128 | No | ||
| 183 | TNFAIP2 | 21144 2100 | 16018 | -0.277 | 0.0148 | No | ||
| 184 | APOA1 | 8614 | 16036 | -0.280 | 0.0186 | No | ||
| 185 | MFNG | 2245 22215 | 16213 | -0.323 | 0.0144 | No | ||
| 186 | TGFB1 | 18332 | 16317 | -0.350 | 0.0147 | No | ||
| 187 | IDE | 4893 | 17053 | -0.611 | -0.0150 | No | ||
| 188 | SCYE1 | 8897 | 17457 | -0.844 | -0.0227 | No | ||
| 189 | FVT1 | 13862 | 17600 | -0.925 | -0.0149 | No | ||
| 190 | LGALS3BP | 20129 | 17602 | -0.927 | 0.0006 | No | ||
| 191 | COPA | 8773 | 17875 | -1.099 | 0.0042 | No | ||
| 192 | NUCB1 | 9494 5201 | 18435 | -2.145 | 0.0098 | No |