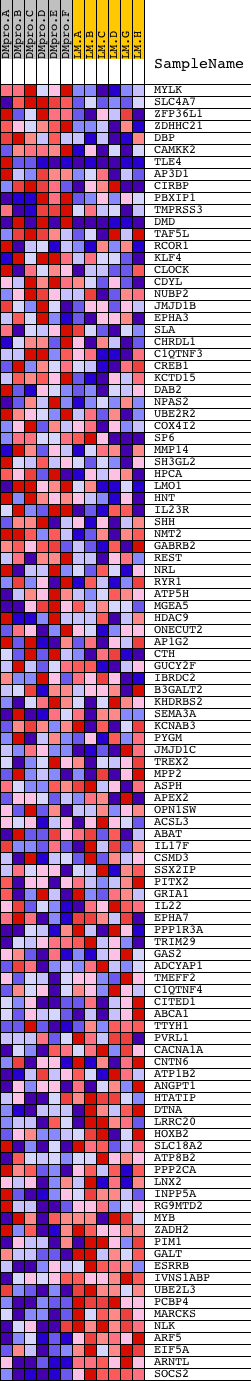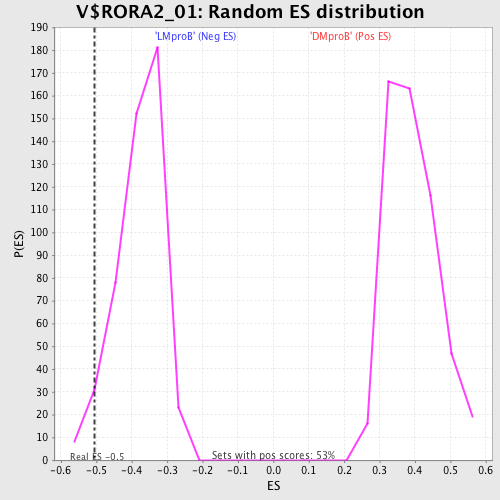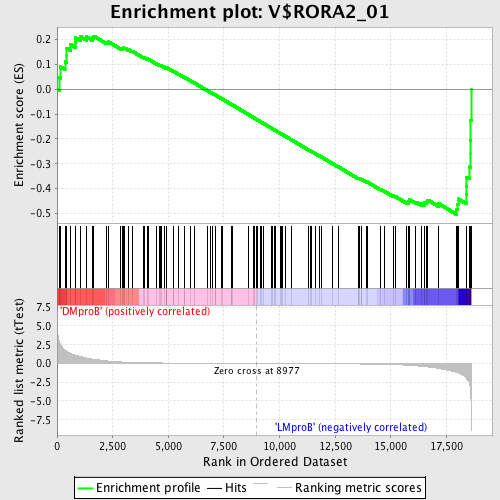
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | V$RORA2_01 |
| Enrichment Score (ES) | -0.50423235 |
| Normalized Enrichment Score (NES) | -1.3360211 |
| Nominal p-value | 0.05073996 |
| FDR q-value | 0.91451055 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MYLK | 22778 4213 | 107 | 2.894 | 0.0469 | No | ||
| 2 | SLC4A7 | 10181 | 160 | 2.535 | 0.0902 | No | ||
| 3 | ZFP36L1 | 4453 4454 | 356 | 1.756 | 0.1116 | No | ||
| 4 | ZDHHC21 | 15854 | 437 | 1.609 | 0.1365 | No | ||
| 5 | DBP | 18242 | 439 | 1.605 | 0.1657 | No | ||
| 6 | CAMKK2 | 9870 | 618 | 1.298 | 0.1796 | No | ||
| 7 | TLE4 | 23720 3697 | 817 | 1.083 | 0.1887 | No | ||
| 8 | AP3D1 | 19690 | 837 | 1.064 | 0.2070 | No | ||
| 9 | CIRBP | 8742 | 1053 | 0.925 | 0.2122 | No | ||
| 10 | PBXIP1 | 6028 | 1319 | 0.731 | 0.2112 | No | ||
| 11 | TMPRSS3 | 23036 | 1583 | 0.587 | 0.2076 | No | ||
| 12 | DMD | 24295 2647 | 1652 | 0.559 | 0.2141 | No | ||
| 13 | TAF5L | 18712 | 2211 | 0.350 | 0.1903 | No | ||
| 14 | RCOR1 | 23998 | 2294 | 0.326 | 0.1918 | No | ||
| 15 | KLF4 | 9230 4962 4963 | 2856 | 0.205 | 0.1653 | No | ||
| 16 | CLOCK | 8749 16507 4530 | 2956 | 0.192 | 0.1634 | No | ||
| 17 | CDYL | 3195 4513 3287 | 2965 | 0.192 | 0.1665 | No | ||
| 18 | NUBP2 | 23082 1513 | 3038 | 0.181 | 0.1659 | No | ||
| 19 | JMJD1B | 23600 | 3190 | 0.162 | 0.1607 | No | ||
| 20 | EPHA3 | 22563 1643 | 3368 | 0.144 | 0.1537 | No | ||
| 21 | SLA | 5449 | 3871 | 0.104 | 0.1285 | No | ||
| 22 | CHRDL1 | 8180 13504 | 3927 | 0.100 | 0.1273 | No | ||
| 23 | C1QTNF3 | 2224 22505 | 4047 | 0.093 | 0.1226 | No | ||
| 24 | CREB1 | 3990 8782 4558 4093 | 4110 | 0.090 | 0.1209 | No | ||
| 25 | KCTD15 | 17864 | 4481 | 0.074 | 0.1023 | No | ||
| 26 | DAB2 | 8835 | 4623 | 0.069 | 0.0959 | No | ||
| 27 | NPAS2 | 5186 | 4628 | 0.069 | 0.0969 | No | ||
| 28 | UBE2R2 | 16240 | 4669 | 0.068 | 0.0960 | No | ||
| 29 | COX4I2 | 14791 | 4805 | 0.063 | 0.0899 | No | ||
| 30 | SP6 | 8178 13678 13679 | 4898 | 0.061 | 0.0860 | No | ||
| 31 | MMP14 | 22020 | 4914 | 0.061 | 0.0863 | No | ||
| 32 | SH3GL2 | 9811 5429 | 4934 | 0.060 | 0.0864 | No | ||
| 33 | HPCA | 9118 | 5220 | 0.053 | 0.0719 | No | ||
| 34 | LMO1 | 17685 | 5230 | 0.053 | 0.0724 | No | ||
| 35 | HNT | 10641 | 5475 | 0.047 | 0.0601 | No | ||
| 36 | IL23R | 17127 | 5720 | 0.042 | 0.0477 | No | ||
| 37 | SHH | 16581 | 5730 | 0.042 | 0.0479 | No | ||
| 38 | NMT2 | 9470 | 5991 | 0.037 | 0.0346 | No | ||
| 39 | GABRB2 | 20922 | 6157 | 0.035 | 0.0263 | No | ||
| 40 | REST | 16818 | 6738 | 0.026 | -0.0046 | No | ||
| 41 | NRL | 9484 | 6873 | 0.024 | -0.0114 | No | ||
| 42 | RYR1 | 17903 17902 9770 | 7000 | 0.022 | -0.0178 | No | ||
| 43 | ATP5H | 12948 | 7096 | 0.021 | -0.0225 | No | ||
| 44 | MGEA5 | 8001 | 7105 | 0.021 | -0.0226 | No | ||
| 45 | HDAC9 | 21084 2131 | 7396 | 0.017 | -0.0380 | No | ||
| 46 | ONECUT2 | 10346 | 7444 | 0.017 | -0.0402 | No | ||
| 47 | AP1G2 | 2677 21827 | 7819 | 0.013 | -0.0602 | No | ||
| 48 | CTH | 1938 15131 | 7834 | 0.012 | -0.0607 | No | ||
| 49 | GUCY2F | 24045 | 7880 | 0.012 | -0.0629 | No | ||
| 50 | IBRDC2 | 5747 | 8612 | 0.004 | -0.1023 | No | ||
| 51 | B3GALT2 | 6498 | 8807 | 0.002 | -0.1128 | No | ||
| 52 | KHDRBS2 | 14284 | 8869 | 0.001 | -0.1161 | No | ||
| 53 | SEMA3A | 16919 | 8963 | 0.000 | -0.1211 | No | ||
| 54 | KCNAB3 | 20824 | 8999 | -0.000 | -0.1230 | No | ||
| 55 | PYGM | 23792 | 9146 | -0.002 | -0.1308 | No | ||
| 56 | JMJD1C | 11531 19996 | 9199 | -0.002 | -0.1336 | No | ||
| 57 | TREX2 | 24137 | 9295 | -0.003 | -0.1387 | No | ||
| 58 | MPP2 | 20209 | 9617 | -0.006 | -0.1559 | No | ||
| 59 | ASPH | 2498 2460 2396 2370 2357 2502 2516 2375 2337 12276 2381 2454 7246 2480 2393 | 9693 | -0.007 | -0.1598 | No | ||
| 60 | APEX2 | 8081 | 9785 | -0.008 | -0.1646 | No | ||
| 61 | OPN1SW | 17196 | 9819 | -0.008 | -0.1662 | No | ||
| 62 | ACSL3 | 13184 | 10041 | -0.010 | -0.1780 | No | ||
| 63 | ABAT | 22864 1713 22669 | 10085 | -0.011 | -0.1801 | No | ||
| 64 | IL17F | 13992 | 10139 | -0.011 | -0.1828 | No | ||
| 65 | CSMD3 | 10735 7938 | 10276 | -0.013 | -0.1899 | No | ||
| 66 | SSX2IP | 13617 | 10550 | -0.016 | -0.2043 | No | ||
| 67 | PITX2 | 15424 1878 | 11281 | -0.024 | -0.2433 | No | ||
| 68 | GRIA1 | 20877 4801 9040 | 11398 | -0.026 | -0.2491 | No | ||
| 69 | IL22 | 11948 | 11453 | -0.026 | -0.2516 | No | ||
| 70 | EPHA7 | 8909 4674 | 11599 | -0.028 | -0.2589 | No | ||
| 71 | PPP1R3A | 17214 | 11802 | -0.031 | -0.2692 | No | ||
| 72 | TRIM29 | 19483 3061 | 11872 | -0.032 | -0.2724 | No | ||
| 73 | GAS2 | 4753 | 12399 | -0.040 | -0.3001 | No | ||
| 74 | ADCYAP1 | 4348 | 12627 | -0.044 | -0.3115 | No | ||
| 75 | TMEFF2 | 7104 12105 | 13532 | -0.065 | -0.3592 | No | ||
| 76 | C1QTNF4 | 14953 | 13558 | -0.065 | -0.3594 | No | ||
| 77 | CITED1 | 24091 | 13592 | -0.066 | -0.3599 | No | ||
| 78 | ABCA1 | 15881 | 13664 | -0.069 | -0.3625 | No | ||
| 79 | TTYH1 | 2496 7176 | 13697 | -0.070 | -0.3630 | No | ||
| 80 | PVRL1 | 3006 19482 12211 7192 | 13915 | -0.078 | -0.3733 | No | ||
| 81 | CACNA1A | 4461 | 13941 | -0.079 | -0.3732 | No | ||
| 82 | CNTN6 | 17346 | 14521 | -0.105 | -0.4026 | No | ||
| 83 | ATP1B2 | 20390 | 14701 | -0.114 | -0.4102 | No | ||
| 84 | ANGPT1 | 8588 2260 22303 | 15105 | -0.144 | -0.4293 | No | ||
| 85 | HTATIP | 3690 | 15203 | -0.154 | -0.4317 | No | ||
| 86 | DTNA | 8870 4647 23611 1991 | 15720 | -0.220 | -0.4556 | No | ||
| 87 | LRRC20 | 20009 | 15787 | -0.232 | -0.4550 | No | ||
| 88 | HOXB2 | 8346 | 15804 | -0.235 | -0.4516 | No | ||
| 89 | SLC18A2 | 5639 | 15829 | -0.239 | -0.4485 | No | ||
| 90 | ATP8B2 | 12055 | 15839 | -0.241 | -0.4446 | No | ||
| 91 | PPP2CA | 20890 | 16118 | -0.299 | -0.4542 | No | ||
| 92 | LNX2 | 16290 | 16360 | -0.361 | -0.4606 | No | ||
| 93 | INPP5A | 10003 | 16511 | -0.399 | -0.4615 | No | ||
| 94 | RG9MTD2 | 8443 | 16528 | -0.405 | -0.4550 | No | ||
| 95 | MYB | 3344 3355 19806 | 16625 | -0.437 | -0.4522 | No | ||
| 96 | ZADH2 | 23501 | 16665 | -0.453 | -0.4461 | No | ||
| 97 | PIM1 | 23308 | 17127 | -0.645 | -0.4593 | No | ||
| 98 | GALT | 16237 2399 | 17960 | -1.173 | -0.4829 | Yes | ||
| 99 | ESRRB | 21196 | 18013 | -1.226 | -0.4634 | Yes | ||
| 100 | IVNS1ABP | 4384 4050 | 18039 | -1.265 | -0.4418 | Yes | ||
| 101 | UBE2L3 | 5818 22647 | 18386 | -1.929 | -0.4254 | Yes | ||
| 102 | PCBP4 | 12235 | 18392 | -1.951 | -0.3902 | Yes | ||
| 103 | MARCKS | 9331 | 18410 | -2.026 | -0.3542 | Yes | ||
| 104 | NLK | 5179 5178 | 18533 | -2.631 | -0.3129 | Yes | ||
| 105 | ARF5 | 8623 | 18565 | -3.022 | -0.2596 | Yes | ||
| 106 | EIF5A | 11345 20379 6590 | 18567 | -3.026 | -0.2047 | Yes | ||
| 107 | ARNTL | 18119 | 18598 | -4.410 | -0.1261 | Yes | ||
| 108 | SOCS2 | 5694 | 18613 | -6.981 | 0.0002 | Yes |