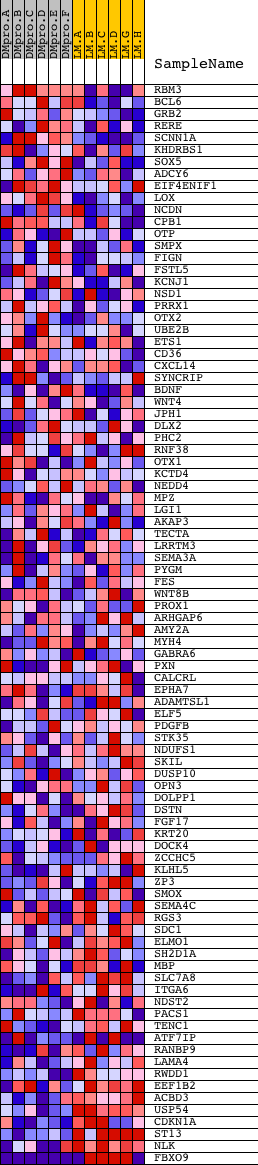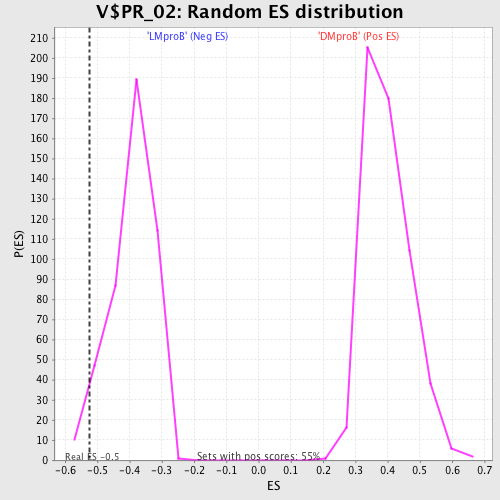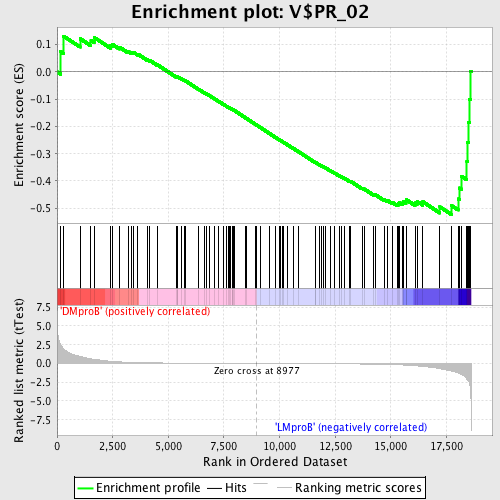
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | V$PR_02 |
| Enrichment Score (ES) | -0.5228441 |
| Normalized Enrichment Score (NES) | -1.3351703 |
| Nominal p-value | 0.03125 |
| FDR q-value | 0.8428569 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RBM3 | 5366 2625 | 162 | 2.521 | 0.0740 | No | ||
| 2 | BCL6 | 22624 | 290 | 1.956 | 0.1313 | No | ||
| 3 | GRB2 | 20149 | 1032 | 0.936 | 0.1220 | No | ||
| 4 | RERE | 12722 | 1522 | 0.619 | 0.1159 | No | ||
| 5 | SCNN1A | 17278 | 1665 | 0.552 | 0.1264 | No | ||
| 6 | KHDRBS1 | 5405 9778 | 2407 | 0.297 | 0.0961 | No | ||
| 7 | SOX5 | 9850 1044 5480 16937 | 2494 | 0.277 | 0.1006 | No | ||
| 8 | ADCY6 | 22139 2283 8551 | 2802 | 0.213 | 0.0910 | No | ||
| 9 | EIF4ENIF1 | 20971 | 3188 | 0.163 | 0.0755 | No | ||
| 10 | LOX | 23438 | 3339 | 0.146 | 0.0722 | No | ||
| 11 | NCDN | 15756 2487 6471 | 3429 | 0.138 | 0.0720 | No | ||
| 12 | CPB1 | 175 | 3621 | 0.121 | 0.0656 | No | ||
| 13 | OTP | 21582 | 4066 | 0.092 | 0.0447 | No | ||
| 14 | SMPX | 24220 | 4164 | 0.088 | 0.0423 | No | ||
| 15 | FIGN | 14575 | 4512 | 0.073 | 0.0260 | No | ||
| 16 | FSTL5 | 10018 | 5361 | 0.050 | -0.0181 | No | ||
| 17 | KCNJ1 | 3121 12110 | 5405 | 0.049 | -0.0188 | No | ||
| 18 | NSD1 | 2134 5197 | 5413 | 0.049 | -0.0176 | No | ||
| 19 | PRRX1 | 9595 5271 | 5575 | 0.045 | -0.0248 | No | ||
| 20 | OTX2 | 9519 | 5718 | 0.042 | -0.0311 | No | ||
| 21 | UBE2B | 10245 | 5750 | 0.042 | -0.0314 | No | ||
| 22 | ETS1 | 10715 6230 3135 | 6350 | 0.031 | -0.0627 | No | ||
| 23 | CD36 | 8712 | 6622 | 0.027 | -0.0765 | No | ||
| 24 | CXCL14 | 21445 7165 | 6731 | 0.026 | -0.0815 | No | ||
| 25 | SYNCRIP | 3078 3035 3107 | 6830 | 0.024 | -0.0859 | No | ||
| 26 | BDNF | 14926 2797 | 7063 | 0.021 | -0.0978 | No | ||
| 27 | WNT4 | 16025 | 7242 | 0.019 | -0.1067 | No | ||
| 28 | JPH1 | 13994 | 7489 | 0.016 | -0.1195 | No | ||
| 29 | DLX2 | 14561 353 | 7617 | 0.015 | -0.1258 | No | ||
| 30 | PHC2 | 16075 | 7707 | 0.014 | -0.1302 | No | ||
| 31 | RNF38 | 7862 2328 13115 | 7731 | 0.014 | -0.1310 | No | ||
| 32 | OTX1 | 20517 | 7814 | 0.013 | -0.1350 | No | ||
| 33 | KCTD4 | 21956 | 7865 | 0.012 | -0.1373 | No | ||
| 34 | NEDD4 | 19384 3101 | 7897 | 0.012 | -0.1386 | No | ||
| 35 | MPZ | 9408 | 7921 | 0.011 | -0.1395 | No | ||
| 36 | LGI1 | 23867 | 7971 | 0.011 | -0.1417 | No | ||
| 37 | AKAP3 | 8566 | 8452 | 0.005 | -0.1675 | No | ||
| 38 | TECTA | 19159 | 8495 | 0.005 | -0.1696 | No | ||
| 39 | LRRTM3 | 19744 | 8922 | 0.001 | -0.1926 | No | ||
| 40 | SEMA3A | 16919 | 8963 | 0.000 | -0.1947 | No | ||
| 41 | PYGM | 23792 | 9146 | -0.002 | -0.2045 | No | ||
| 42 | FES | 17779 3949 | 9550 | -0.006 | -0.2260 | No | ||
| 43 | WNT8B | 23841 | 9798 | -0.008 | -0.2391 | No | ||
| 44 | PROX1 | 9623 | 10005 | -0.010 | -0.2499 | No | ||
| 45 | ARHGAP6 | 24207 | 10039 | -0.010 | -0.2514 | No | ||
| 46 | AMY2A | 8587 | 10122 | -0.011 | -0.2554 | No | ||
| 47 | MYH4 | 20837 | 10179 | -0.012 | -0.2581 | No | ||
| 48 | GABRA6 | 20491 | 10373 | -0.014 | -0.2680 | No | ||
| 49 | PXN | 5339 3573 | 10630 | -0.017 | -0.2813 | No | ||
| 50 | CALCRL | 12041 | 10863 | -0.019 | -0.2932 | No | ||
| 51 | EPHA7 | 8909 4674 | 11599 | -0.028 | -0.3319 | No | ||
| 52 | ADAMTSL1 | 16181 | 11605 | -0.029 | -0.3313 | No | ||
| 53 | ELF5 | 14936 2913 | 11796 | -0.031 | -0.3405 | No | ||
| 54 | PDGFB | 22199 2311 | 11899 | -0.033 | -0.3449 | No | ||
| 55 | STK35 | 14851 | 11969 | -0.033 | -0.3476 | No | ||
| 56 | NDUFS1 | 13940 | 12057 | -0.035 | -0.3511 | No | ||
| 57 | SKIL | 15617 | 12286 | -0.038 | -0.3622 | No | ||
| 58 | DUSP10 | 4003 14016 | 12469 | -0.041 | -0.3706 | No | ||
| 59 | OPN3 | 13740 | 12698 | -0.045 | -0.3815 | No | ||
| 60 | DOLPP1 | 2682 15051 | 12790 | -0.047 | -0.3848 | No | ||
| 61 | DSTN | 14823 | 12909 | -0.049 | -0.3896 | No | ||
| 62 | FGF17 | 21757 | 13164 | -0.054 | -0.4015 | No | ||
| 63 | KRT20 | 12410 | 13196 | -0.055 | -0.4014 | No | ||
| 64 | DOCK4 | 10705 | 13711 | -0.070 | -0.4268 | No | ||
| 65 | ZCCHC5 | 24081 | 13802 | -0.074 | -0.4293 | No | ||
| 66 | KLHL5 | 3631 7753 | 14235 | -0.091 | -0.4496 | No | ||
| 67 | ZP3 | 3509 | 14296 | -0.093 | -0.4498 | No | ||
| 68 | SMOX | 10455 2940 2737 2952 | 14718 | -0.115 | -0.4687 | No | ||
| 69 | SEMA4C | 9799 | 14863 | -0.125 | -0.4724 | No | ||
| 70 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 15065 | -0.142 | -0.4786 | No | ||
| 71 | SDC1 | 5560 9953 | 15305 | -0.164 | -0.4861 | No | ||
| 72 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 15345 | -0.168 | -0.4827 | No | ||
| 73 | SH2D1A | 9810 | 15381 | -0.172 | -0.4789 | No | ||
| 74 | MBP | 23502 | 15536 | -0.194 | -0.4809 | No | ||
| 75 | SLC7A8 | 3611 21833 | 15553 | -0.197 | -0.4753 | No | ||
| 76 | ITGA6 | 4930 | 15683 | -0.215 | -0.4752 | No | ||
| 77 | NDST2 | 21906 | 15694 | -0.216 | -0.4687 | No | ||
| 78 | PACS1 | 23975 | 16089 | -0.291 | -0.4804 | No | ||
| 79 | TENC1 | 22349 9927 | 16180 | -0.314 | -0.4749 | No | ||
| 80 | ATF7IP | 12028 13038 | 16441 | -0.384 | -0.4764 | No | ||
| 81 | RANBP9 | 21480 | 17204 | -0.684 | -0.4950 | Yes | ||
| 82 | LAMA4 | 20054 | 17720 | -1.000 | -0.4900 | Yes | ||
| 83 | RWDD1 | 7306 | 18025 | -1.236 | -0.4659 | Yes | ||
| 84 | EEF1B2 | 4131 12063 | 18086 | -1.325 | -0.4256 | Yes | ||
| 85 | ACBD3 | 14025 | 18170 | -1.461 | -0.3822 | Yes | ||
| 86 | USP54 | 21910 | 18419 | -2.080 | -0.3273 | Yes | ||
| 87 | CDKN1A | 4511 8729 | 18443 | -2.164 | -0.2575 | Yes | ||
| 88 | ST13 | 12865 | 18484 | -2.298 | -0.1843 | Yes | ||
| 89 | NLK | 5179 5178 | 18533 | -2.631 | -0.1005 | Yes | ||
| 90 | FBXO9 | 19058 3146 | 18575 | -3.199 | 0.0022 | Yes |