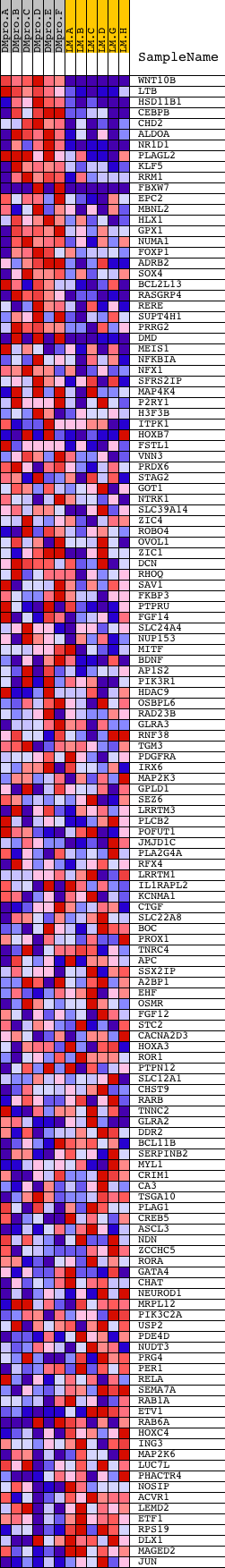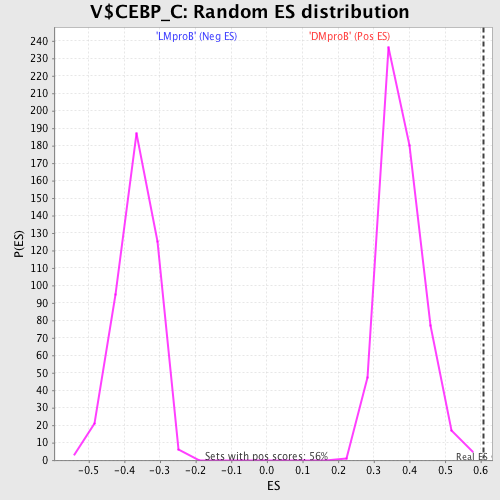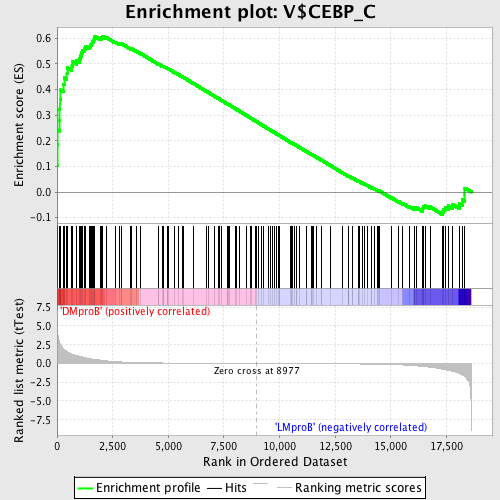
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | DMproB |
| GeneSet | V$CEBP_C |
| Enrichment Score (ES) | 0.60819346 |
| Normalized Enrichment Score (NES) | 1.6053647 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.51470304 |
| FWER p-Value | 0.351 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | WNT10B | 5874 | 0 | 6.985 | 0.1032 | Yes | ||
| 2 | LTB | 23259 | 6 | 5.641 | 0.1863 | Yes | ||
| 3 | HSD11B1 | 9125 4019 | 34 | 3.879 | 0.2421 | Yes | ||
| 4 | CEBPB | 8733 | 115 | 2.822 | 0.2795 | Yes | ||
| 5 | CHD2 | 10847 | 119 | 2.805 | 0.3208 | Yes | ||
| 6 | ALDOA | 8572 | 129 | 2.714 | 0.3604 | Yes | ||
| 7 | NR1D1 | 4650 20258 949 | 146 | 2.616 | 0.3982 | Yes | ||
| 8 | PLAGL2 | 7055 | 280 | 1.985 | 0.4203 | Yes | ||
| 9 | KLF5 | 4456 | 328 | 1.857 | 0.4452 | Yes | ||
| 10 | RRM1 | 18163 | 443 | 1.598 | 0.4626 | Yes | ||
| 11 | FBXW7 | 1805 11928 | 450 | 1.581 | 0.4857 | Yes | ||
| 12 | EPC2 | 15015 | 668 | 1.235 | 0.4922 | Yes | ||
| 13 | MBNL2 | 21932 | 702 | 1.192 | 0.5080 | Yes | ||
| 14 | HLX1 | 9093 | 876 | 1.034 | 0.5139 | Yes | ||
| 15 | GPX1 | 19310 | 1017 | 0.947 | 0.5203 | Yes | ||
| 16 | NUMA1 | 18167 | 1046 | 0.927 | 0.5325 | Yes | ||
| 17 | FOXP1 | 4242 | 1101 | 0.886 | 0.5427 | Yes | ||
| 18 | ADRB2 | 23422 | 1144 | 0.855 | 0.5530 | Yes | ||
| 19 | SOX4 | 5479 | 1229 | 0.790 | 0.5601 | Yes | ||
| 20 | BCL2L13 | 17302 | 1281 | 0.753 | 0.5685 | Yes | ||
| 21 | RASGRP4 | 6119 | 1468 | 0.642 | 0.5679 | Yes | ||
| 22 | RERE | 12722 | 1522 | 0.619 | 0.5742 | Yes | ||
| 23 | SUPT4H1 | 9938 | 1565 | 0.594 | 0.5807 | Yes | ||
| 24 | PRRG2 | 17841 | 1572 | 0.592 | 0.5891 | Yes | ||
| 25 | DMD | 24295 2647 | 1652 | 0.559 | 0.5931 | Yes | ||
| 26 | MEIS1 | 20524 | 1660 | 0.553 | 0.6009 | Yes | ||
| 27 | NFKBIA | 21065 | 1694 | 0.538 | 0.6071 | Yes | ||
| 28 | NFX1 | 2475 | 1933 | 0.444 | 0.6007 | Yes | ||
| 29 | SFRS2IP | 7794 13009 | 2013 | 0.407 | 0.6025 | Yes | ||
| 30 | MAP4K4 | 11241 | 2019 | 0.406 | 0.6082 | Yes | ||
| 31 | P2RY1 | 15581 | 2198 | 0.353 | 0.6038 | No | ||
| 32 | H3F3B | 4839 | 2610 | 0.250 | 0.5852 | No | ||
| 33 | ITPK1 | 10150 | 2817 | 0.211 | 0.5772 | No | ||
| 34 | HOXB7 | 666 9110 | 2877 | 0.202 | 0.5770 | No | ||
| 35 | FSTL1 | 22765 | 2910 | 0.198 | 0.5782 | No | ||
| 36 | VNN3 | 20072 3422 | 3295 | 0.151 | 0.5596 | No | ||
| 37 | PRDX6 | 4389 8596 | 3335 | 0.147 | 0.5597 | No | ||
| 38 | STAG2 | 5521 | 3550 | 0.126 | 0.5500 | No | ||
| 39 | GOT1 | 23670 | 3765 | 0.111 | 0.5400 | No | ||
| 40 | NTRK1 | 15299 | 4547 | 0.072 | 0.4988 | No | ||
| 41 | SLC39A14 | 3517 5611 21762 | 4552 | 0.072 | 0.4997 | No | ||
| 42 | ZIC4 | 5992 | 4746 | 0.065 | 0.4902 | No | ||
| 43 | ROBO4 | 7895 3064 | 4773 | 0.065 | 0.4897 | No | ||
| 44 | OVOL1 | 23983 | 4786 | 0.064 | 0.4900 | No | ||
| 45 | ZIC1 | 19040 | 4972 | 0.059 | 0.4809 | No | ||
| 46 | DCN | 19897 4601 8840 3366 | 4981 | 0.059 | 0.4813 | No | ||
| 47 | RHOQ | 23142 | 4991 | 0.059 | 0.4817 | No | ||
| 48 | SAV1 | 21047 | 5292 | 0.051 | 0.4662 | No | ||
| 49 | FKBP3 | 21056 | 5472 | 0.047 | 0.4572 | No | ||
| 50 | PTPRU | 15738 | 5638 | 0.044 | 0.4490 | No | ||
| 51 | FGF14 | 21716 3005 3463 | 5686 | 0.043 | 0.4471 | No | ||
| 52 | SLC24A4 | 6223 | 6146 | 0.035 | 0.4227 | No | ||
| 53 | NUP153 | 21474 | 6702 | 0.026 | 0.3931 | No | ||
| 54 | MITF | 17349 | 6805 | 0.025 | 0.3879 | No | ||
| 55 | BDNF | 14926 2797 | 7063 | 0.021 | 0.3743 | No | ||
| 56 | AP1S2 | 8423 4228 | 7235 | 0.019 | 0.3654 | No | ||
| 57 | PIK3R1 | 3170 | 7282 | 0.019 | 0.3631 | No | ||
| 58 | HDAC9 | 21084 2131 | 7396 | 0.017 | 0.3573 | No | ||
| 59 | OSBPL6 | 8277 | 7672 | 0.014 | 0.3426 | No | ||
| 60 | RAD23B | 5349 | 7673 | 0.014 | 0.3428 | No | ||
| 61 | GLRA3 | 18617 | 7691 | 0.014 | 0.3421 | No | ||
| 62 | RNF38 | 7862 2328 13115 | 7731 | 0.014 | 0.3402 | No | ||
| 63 | TGM3 | 10168 | 8033 | 0.010 | 0.3241 | No | ||
| 64 | PDGFRA | 16824 | 8040 | 0.010 | 0.3239 | No | ||
| 65 | IRX6 | 7225 | 8187 | 0.008 | 0.3161 | No | ||
| 66 | MAP2K3 | 20856 | 8518 | 0.004 | 0.2983 | No | ||
| 67 | GPLD1 | 21691 3272 | 8712 | 0.003 | 0.2879 | No | ||
| 68 | SEZ6 | 20764 | 8728 | 0.003 | 0.2871 | No | ||
| 69 | LRRTM3 | 19744 | 8922 | 0.001 | 0.2767 | No | ||
| 70 | PLCB2 | 5262 | 8945 | 0.000 | 0.2755 | No | ||
| 71 | POFUT1 | 4697 | 9033 | -0.001 | 0.2708 | No | ||
| 72 | JMJD1C | 11531 19996 | 9199 | -0.002 | 0.2619 | No | ||
| 73 | PLA2G4A | 13809 | 9258 | -0.003 | 0.2588 | No | ||
| 74 | RFX4 | 7721 3410 | 9495 | -0.005 | 0.2461 | No | ||
| 75 | LRRTM1 | 17408 | 9602 | -0.006 | 0.2405 | No | ||
| 76 | IL1RAPL2 | 24241 | 9666 | -0.007 | 0.2372 | No | ||
| 77 | KCNMA1 | 4947 | 9754 | -0.008 | 0.2326 | No | ||
| 78 | CTGF | 20068 | 9870 | -0.009 | 0.2265 | No | ||
| 79 | SLC22A8 | 23944 | 9942 | -0.009 | 0.2228 | No | ||
| 80 | BOC | 22587 | 9995 | -0.010 | 0.2201 | No | ||
| 81 | PROX1 | 9623 | 10005 | -0.010 | 0.2198 | No | ||
| 82 | TNRC4 | 15514 | 10503 | -0.015 | 0.1931 | No | ||
| 83 | APC | 4396 2022 | 10515 | -0.015 | 0.1927 | No | ||
| 84 | SSX2IP | 13617 | 10550 | -0.016 | 0.1911 | No | ||
| 85 | A2BP1 | 11212 1652 1689 | 10578 | -0.016 | 0.1899 | No | ||
| 86 | EHF | 4657 | 10672 | -0.017 | 0.1851 | No | ||
| 87 | OSMR | 22333 | 10751 | -0.018 | 0.1812 | No | ||
| 88 | FGF12 | 1723 22621 | 10779 | -0.018 | 0.1800 | No | ||
| 89 | STC2 | 5526 | 10885 | -0.020 | 0.1746 | No | ||
| 90 | CACNA2D3 | 8677 | 11201 | -0.023 | 0.1579 | No | ||
| 91 | HOXA3 | 9108 1075 | 11208 | -0.024 | 0.1579 | No | ||
| 92 | ROR1 | 6472 | 11440 | -0.026 | 0.1458 | No | ||
| 93 | PTPN12 | 16597 3502 3586 3665 | 11449 | -0.026 | 0.1457 | No | ||
| 94 | SLC12A1 | 14874 2772 2943 | 11470 | -0.027 | 0.1451 | No | ||
| 95 | CHST9 | 7727 | 11509 | -0.027 | 0.1434 | No | ||
| 96 | RARB | 3011 21915 | 11676 | -0.030 | 0.1349 | No | ||
| 97 | TNNC2 | 14343 | 11868 | -0.032 | 0.1250 | No | ||
| 98 | GLRA2 | 24008 | 11902 | -0.033 | 0.1237 | No | ||
| 99 | DDR2 | 13767 | 12287 | -0.038 | 0.1035 | No | ||
| 100 | BCL11B | 7190 | 12823 | -0.047 | 0.0752 | No | ||
| 101 | SERPINB2 | 14161 | 13092 | -0.053 | 0.0615 | No | ||
| 102 | MYL1 | 4015 | 13102 | -0.053 | 0.0618 | No | ||
| 103 | CRIM1 | 403 | 13113 | -0.053 | 0.0620 | No | ||
| 104 | CA3 | 4476 8690 | 13259 | -0.057 | 0.0550 | No | ||
| 105 | TSGA10 | 4123 13975 5587 | 13287 | -0.058 | 0.0544 | No | ||
| 106 | PLAG1 | 7148 15949 12161 | 13539 | -0.065 | 0.0418 | No | ||
| 107 | CREB5 | 10551 | 13540 | -0.065 | 0.0428 | No | ||
| 108 | ASCL3 | 17681 | 13587 | -0.066 | 0.0412 | No | ||
| 109 | NDN | 18222 | 13708 | -0.070 | 0.0358 | No | ||
| 110 | ZCCHC5 | 24081 | 13802 | -0.074 | 0.0318 | No | ||
| 111 | RORA | 3019 5388 | 13938 | -0.079 | 0.0257 | No | ||
| 112 | GATA4 | 21792 4755 | 14138 | -0.087 | 0.0162 | No | ||
| 113 | CHAT | 21882 | 14144 | -0.087 | 0.0172 | No | ||
| 114 | NEUROD1 | 14550 | 14275 | -0.092 | 0.0116 | No | ||
| 115 | MRPL12 | 12090 | 14403 | -0.098 | 0.0061 | No | ||
| 116 | PIK3C2A | 17667 | 14460 | -0.101 | 0.0046 | No | ||
| 117 | USP2 | 19480 3052 3043 | 14497 | -0.103 | 0.0042 | No | ||
| 118 | PDE4D | 10722 6235 | 15012 | -0.136 | -0.0216 | No | ||
| 119 | NUDT3 | 23056 | 15351 | -0.169 | -0.0374 | No | ||
| 120 | PRG4 | 13592 4036 | 15531 | -0.193 | -0.0442 | No | ||
| 121 | PER1 | 20827 | 15850 | -0.243 | -0.0579 | No | ||
| 122 | RELA | 23783 | 16057 | -0.286 | -0.0648 | No | ||
| 123 | SEMA7A | 19426 9803 | 16067 | -0.288 | -0.0610 | No | ||
| 124 | RAB1A | 20942 | 16171 | -0.311 | -0.0620 | No | ||
| 125 | ETV1 | 4688 8925 8924 | 16408 | -0.374 | -0.0692 | No | ||
| 126 | RAB6A | 18173 3905 | 16412 | -0.376 | -0.0638 | No | ||
| 127 | HOXC4 | 4864 | 16454 | -0.386 | -0.0604 | No | ||
| 128 | ING3 | 17514 1019 | 16481 | -0.391 | -0.0560 | No | ||
| 129 | MAP2K6 | 20614 1414 | 16564 | -0.417 | -0.0543 | No | ||
| 130 | LUC7L | 47 1520 | 16765 | -0.495 | -0.0578 | No | ||
| 131 | PHACTR4 | 4137 | 17318 | -0.749 | -0.0766 | No | ||
| 132 | NOSIP | 18254 3847 | 17374 | -0.783 | -0.0680 | No | ||
| 133 | ACVR1 | 4334 | 17437 | -0.829 | -0.0591 | No | ||
| 134 | LEMD2 | 5893 | 17579 | -0.907 | -0.0533 | No | ||
| 135 | ETF1 | 23467 | 17789 | -1.041 | -0.0493 | No | ||
| 136 | RPS19 | 5398 | 18077 | -1.312 | -0.0454 | No | ||
| 137 | DLX1 | 14988 | 18212 | -1.546 | -0.0298 | No | ||
| 138 | MAGED2 | 24034 | 18328 | -1.742 | -0.0103 | No | ||
| 139 | JUN | 15832 | 18330 | -1.748 | 0.0155 | No |