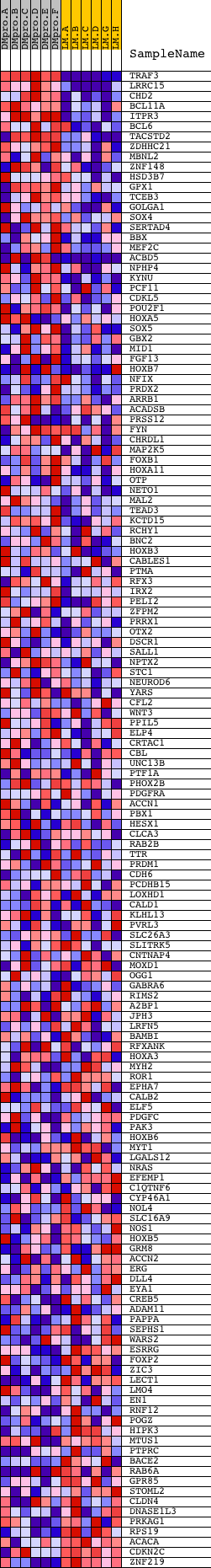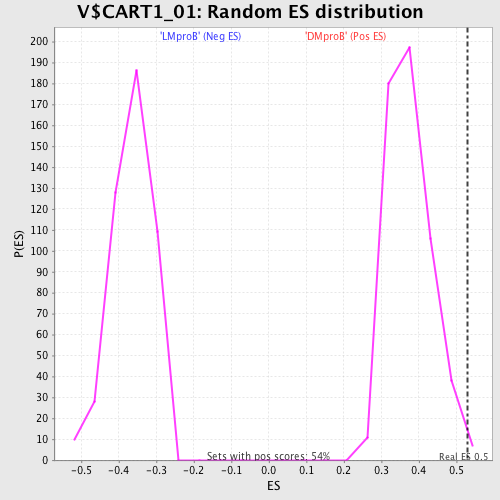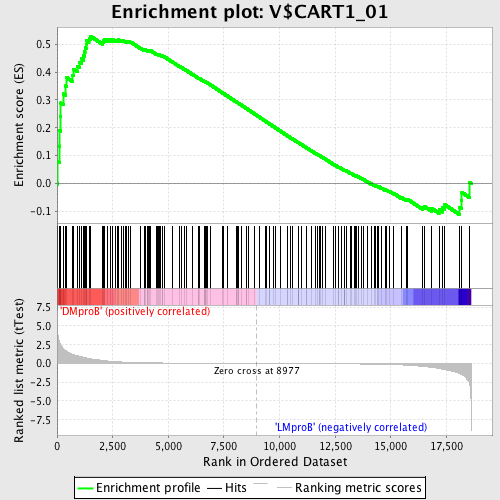
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | DMproB |
| GeneSet | V$CART1_01 |
| Enrichment Score (ES) | 0.52860093 |
| Normalized Enrichment Score (NES) | 1.4084847 |
| Nominal p-value | 0.0074211503 |
| FDR q-value | 0.40576127 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TRAF3 | 21147 | 30 | 4.000 | 0.0782 | Yes | ||
| 2 | LRRC15 | 22617 | 95 | 3.011 | 0.1348 | Yes | ||
| 3 | CHD2 | 10847 | 119 | 2.805 | 0.1896 | Yes | ||
| 4 | BCL11A | 4691 | 147 | 2.605 | 0.2401 | Yes | ||
| 5 | ITPR3 | 9195 | 166 | 2.502 | 0.2890 | Yes | ||
| 6 | BCL6 | 22624 | 290 | 1.956 | 0.3214 | Yes | ||
| 7 | TACSTD2 | 17126 | 386 | 1.692 | 0.3501 | Yes | ||
| 8 | ZDHHC21 | 15854 | 437 | 1.609 | 0.3795 | Yes | ||
| 9 | MBNL2 | 21932 | 702 | 1.192 | 0.3889 | Yes | ||
| 10 | ZNF148 | 5949 | 749 | 1.147 | 0.4094 | Yes | ||
| 11 | HSD3B7 | 4158 | 910 | 1.013 | 0.4209 | Yes | ||
| 12 | GPX1 | 19310 | 1017 | 0.947 | 0.4341 | Yes | ||
| 13 | TCEB3 | 15707 | 1094 | 0.895 | 0.4478 | Yes | ||
| 14 | GOLGA1 | 14595 | 1195 | 0.816 | 0.4587 | Yes | ||
| 15 | SOX4 | 5479 | 1229 | 0.790 | 0.4727 | Yes | ||
| 16 | SERTAD4 | 13715 | 1256 | 0.775 | 0.4867 | Yes | ||
| 17 | BBX | 7685 | 1311 | 0.733 | 0.4984 | Yes | ||
| 18 | MEF2C | 3204 9378 | 1329 | 0.725 | 0.5120 | Yes | ||
| 19 | ACBD5 | 15105 2751 | 1434 | 0.667 | 0.5196 | Yes | ||
| 20 | NPHP4 | 15978 | 1500 | 0.626 | 0.5286 | Yes | ||
| 21 | KYNU | 15016 | 2050 | 0.397 | 0.5068 | No | ||
| 22 | PCF11 | 121 | 2087 | 0.385 | 0.5125 | No | ||
| 23 | CDKL5 | 24019 | 2146 | 0.368 | 0.5167 | No | ||
| 24 | POU2F1 | 5275 3989 4065 4010 | 2284 | 0.329 | 0.5159 | No | ||
| 25 | HOXA5 | 17149 | 2380 | 0.304 | 0.5168 | No | ||
| 26 | SOX5 | 9850 1044 5480 16937 | 2494 | 0.277 | 0.5162 | No | ||
| 27 | GBX2 | 13887 | 2627 | 0.247 | 0.5140 | No | ||
| 28 | MID1 | 5097 5098 | 2729 | 0.226 | 0.5130 | No | ||
| 29 | FGF13 | 4720 8963 2627 | 2765 | 0.219 | 0.5155 | No | ||
| 30 | HOXB7 | 666 9110 | 2877 | 0.202 | 0.5135 | No | ||
| 31 | NFIX | 9458 | 2978 | 0.190 | 0.5119 | No | ||
| 32 | PRDX2 | 3871 18542 5705 | 3075 | 0.176 | 0.5102 | No | ||
| 33 | ARRB1 | 3890 8465 18178 4276 | 3130 | 0.170 | 0.5107 | No | ||
| 34 | ACADSB | 7358 | 3213 | 0.160 | 0.5095 | No | ||
| 35 | PRSS12 | 1920 15428 | 3305 | 0.150 | 0.5075 | No | ||
| 36 | FYN | 3375 3395 20052 | 3756 | 0.112 | 0.4854 | No | ||
| 37 | CHRDL1 | 8180 13504 | 3927 | 0.100 | 0.4782 | No | ||
| 38 | MAP2K5 | 19088 | 3943 | 0.099 | 0.4794 | No | ||
| 39 | FOXB1 | 12254 19069 | 3967 | 0.098 | 0.4801 | No | ||
| 40 | HOXA11 | 17146 | 4065 | 0.092 | 0.4767 | No | ||
| 41 | OTP | 21582 | 4066 | 0.092 | 0.4785 | No | ||
| 42 | NETO1 | 6372 1970 | 4124 | 0.090 | 0.4772 | No | ||
| 43 | MAL2 | 22476 | 4167 | 0.087 | 0.4767 | No | ||
| 44 | TEAD3 | 23053 | 4178 | 0.087 | 0.4779 | No | ||
| 45 | KCTD15 | 17864 | 4481 | 0.074 | 0.4630 | No | ||
| 46 | RCHY1 | 12646 | 4499 | 0.074 | 0.4636 | No | ||
| 47 | BNC2 | 15850 | 4563 | 0.072 | 0.4616 | No | ||
| 48 | HOXB3 | 20685 | 4586 | 0.071 | 0.4618 | No | ||
| 49 | CABLES1 | 23621 12249 | 4630 | 0.069 | 0.4609 | No | ||
| 50 | PTMA | 9657 | 4722 | 0.066 | 0.4572 | No | ||
| 51 | RFX3 | 9722 5377 3712 3762 | 4726 | 0.066 | 0.4584 | No | ||
| 52 | IRX2 | 4927 | 4813 | 0.063 | 0.4550 | No | ||
| 53 | PELI2 | 8217 | 5180 | 0.054 | 0.4363 | No | ||
| 54 | ZFPM2 | 22483 | 5479 | 0.047 | 0.4211 | No | ||
| 55 | PRRX1 | 9595 5271 | 5575 | 0.045 | 0.4168 | No | ||
| 56 | OTX2 | 9519 | 5718 | 0.042 | 0.4100 | No | ||
| 57 | DSCR1 | 22536 1633 | 5821 | 0.041 | 0.4053 | No | ||
| 58 | SALL1 | 18796 12207 | 6090 | 0.036 | 0.3915 | No | ||
| 59 | NPTX2 | 16630 | 6367 | 0.031 | 0.3772 | No | ||
| 60 | STC1 | 21974 | 6375 | 0.031 | 0.3774 | No | ||
| 61 | NEUROD6 | 17138 | 6384 | 0.031 | 0.3776 | No | ||
| 62 | YARS | 16071 | 6617 | 0.027 | 0.3655 | No | ||
| 63 | CFL2 | 2155 21069 | 6618 | 0.027 | 0.3661 | No | ||
| 64 | WNT3 | 20635 | 6648 | 0.027 | 0.3650 | No | ||
| 65 | PPIL5 | 21255 | 6691 | 0.026 | 0.3633 | No | ||
| 66 | ELP4 | 8094 | 6704 | 0.026 | 0.3632 | No | ||
| 67 | CRTAC1 | 23673 | 6749 | 0.025 | 0.3613 | No | ||
| 68 | CBL | 19154 | 6906 | 0.023 | 0.3533 | No | ||
| 69 | UNC13B | 10255 | 7454 | 0.017 | 0.3240 | No | ||
| 70 | PTF1A | 15110 | 7462 | 0.017 | 0.3240 | No | ||
| 71 | PHOX2B | 16524 | 7669 | 0.014 | 0.3131 | No | ||
| 72 | PDGFRA | 16824 | 8040 | 0.010 | 0.2933 | No | ||
| 73 | ACCN1 | 20327 1194 | 8049 | 0.010 | 0.2930 | No | ||
| 74 | PBX1 | 5224 | 8124 | 0.009 | 0.2892 | No | ||
| 75 | HESX1 | 22070 | 8143 | 0.009 | 0.2884 | No | ||
| 76 | CLCA3 | 6224 | 8150 | 0.009 | 0.2883 | No | ||
| 77 | RAB2B | 21842 | 8305 | 0.007 | 0.2801 | No | ||
| 78 | TTR | 23615 | 8503 | 0.005 | 0.2695 | No | ||
| 79 | PRDM1 | 19775 3337 | 8527 | 0.004 | 0.2683 | No | ||
| 80 | CDH6 | 22320 | 8601 | 0.004 | 0.2645 | No | ||
| 81 | PCDHB15 | 13560 | 8859 | 0.001 | 0.2506 | No | ||
| 82 | LOXHD1 | 23507 | 9074 | -0.001 | 0.2390 | No | ||
| 83 | CALD1 | 4273 8463 | 9344 | -0.004 | 0.2245 | No | ||
| 84 | KLHL13 | 12536 | 9392 | -0.004 | 0.2221 | No | ||
| 85 | PVRL3 | 1674 22579 1675 | 9526 | -0.006 | 0.2150 | No | ||
| 86 | SLC26A3 | 21298 | 9549 | -0.006 | 0.2139 | No | ||
| 87 | SLITRK5 | 21939 | 9712 | -0.007 | 0.2053 | No | ||
| 88 | CNTNAP4 | 18462 3857 | 9799 | -0.008 | 0.2008 | No | ||
| 89 | MOXD1 | 20069 | 10061 | -0.011 | 0.1868 | No | ||
| 90 | OGG1 | 17331 | 10363 | -0.014 | 0.1708 | No | ||
| 91 | GABRA6 | 20491 | 10373 | -0.014 | 0.1706 | No | ||
| 92 | RIMS2 | 4370 | 10510 | -0.015 | 0.1635 | No | ||
| 93 | A2BP1 | 11212 1652 1689 | 10578 | -0.016 | 0.1602 | No | ||
| 94 | JPH3 | 18441 | 10849 | -0.019 | 0.1460 | No | ||
| 95 | LRFN5 | 6213 | 10853 | -0.019 | 0.1462 | No | ||
| 96 | BAMBI | 23629 | 10857 | -0.019 | 0.1464 | No | ||
| 97 | RFXANK | 5378 | 11002 | -0.021 | 0.1391 | No | ||
| 98 | HOXA3 | 9108 1075 | 11208 | -0.024 | 0.1284 | No | ||
| 99 | MYH2 | 20838 | 11231 | -0.024 | 0.1277 | No | ||
| 100 | ROR1 | 6472 | 11440 | -0.026 | 0.1170 | No | ||
| 101 | EPHA7 | 8909 4674 | 11599 | -0.028 | 0.1090 | No | ||
| 102 | CALB2 | 8680 18748 | 11698 | -0.030 | 0.1043 | No | ||
| 103 | ELF5 | 14936 2913 | 11796 | -0.031 | 0.0996 | No | ||
| 104 | PDGFC | 7048 | 11806 | -0.031 | 0.0998 | No | ||
| 105 | PAK3 | 9528 | 11842 | -0.032 | 0.0985 | No | ||
| 106 | HOXB6 | 20688 | 11919 | -0.033 | 0.0951 | No | ||
| 107 | MYT1 | 36 | 12083 | -0.035 | 0.0869 | No | ||
| 108 | LGALS12 | 12076 | 12411 | -0.040 | 0.0700 | No | ||
| 109 | NRAS | 5191 | 12493 | -0.042 | 0.0665 | No | ||
| 110 | EFEMP1 | 20937 | 12657 | -0.044 | 0.0585 | No | ||
| 111 | C1QTNF6 | 13063 | 12664 | -0.044 | 0.0591 | No | ||
| 112 | CYP46A1 | 21158 | 12789 | -0.047 | 0.0533 | No | ||
| 113 | NOL4 | 2028 11432 | 12911 | -0.049 | 0.0477 | No | ||
| 114 | SLC16A9 | 19993 | 12928 | -0.049 | 0.0479 | No | ||
| 115 | NOS1 | 9474 | 12933 | -0.049 | 0.0486 | No | ||
| 116 | HOXB5 | 20687 | 12998 | -0.051 | 0.0462 | No | ||
| 117 | GRM8 | 4805 | 13203 | -0.055 | 0.0362 | No | ||
| 118 | ACCN2 | 22363 | 13212 | -0.056 | 0.0369 | No | ||
| 119 | ERG | 1686 8915 | 13385 | -0.060 | 0.0288 | No | ||
| 120 | DLL4 | 7040 | 13421 | -0.061 | 0.0281 | No | ||
| 121 | EYA1 | 4695 4061 | 13446 | -0.062 | 0.0281 | No | ||
| 122 | CREB5 | 10551 | 13540 | -0.065 | 0.0243 | No | ||
| 123 | ADAM11 | 4340 1432 | 13674 | -0.069 | 0.0185 | No | ||
| 124 | PAPPA | 16190 | 13774 | -0.073 | 0.0146 | No | ||
| 125 | SEPHS1 | 15125 | 13966 | -0.080 | 0.0058 | No | ||
| 126 | WARS2 | 12884 7688 | 14109 | -0.086 | -0.0001 | No | ||
| 127 | ESRRG | 6447 | 14264 | -0.092 | -0.0066 | No | ||
| 128 | FOXP2 | 4312 17523 8523 | 14322 | -0.095 | -0.0078 | No | ||
| 129 | ZIC3 | 10430 | 14395 | -0.098 | -0.0098 | No | ||
| 130 | LECT1 | 21740 | 14451 | -0.101 | -0.0108 | No | ||
| 131 | LMO4 | 15151 | 14600 | -0.109 | -0.0166 | No | ||
| 132 | EN1 | 387 14155 | 14746 | -0.117 | -0.0221 | No | ||
| 133 | RNF12 | 5384 | 14805 | -0.121 | -0.0228 | No | ||
| 134 | POGZ | 6031 1911 | 14930 | -0.130 | -0.0270 | No | ||
| 135 | HIPK3 | 14504 | 15122 | -0.146 | -0.0344 | No | ||
| 136 | MTUS1 | 4162 158 | 15459 | -0.183 | -0.0489 | No | ||
| 137 | PTPRC | 5327 9662 | 15705 | -0.217 | -0.0579 | No | ||
| 138 | BACE2 | 1738 22687 1695 | 15749 | -0.225 | -0.0557 | No | ||
| 139 | RAB6A | 18173 3905 | 16412 | -0.376 | -0.0840 | No | ||
| 140 | GPR85 | 7227 1048 | 16501 | -0.396 | -0.0809 | No | ||
| 141 | STOML2 | 15902 | 16834 | -0.529 | -0.0883 | No | ||
| 142 | CLDN4 | 8747 | 17169 | -0.663 | -0.0932 | No | ||
| 143 | DNASE1L3 | 21925 | 17330 | -0.757 | -0.0867 | No | ||
| 144 | PRKAG1 | 22135 | 17428 | -0.824 | -0.0755 | No | ||
| 145 | RPS19 | 5398 | 18077 | -1.312 | -0.0844 | No | ||
| 146 | ACACA | 309 4209 | 18173 | -1.470 | -0.0602 | No | ||
| 147 | CDKN2C | 15806 2321 | 18193 | -1.505 | -0.0312 | No | ||
| 148 | ZNF219 | 12835 | 18537 | -2.709 | 0.0043 | No |