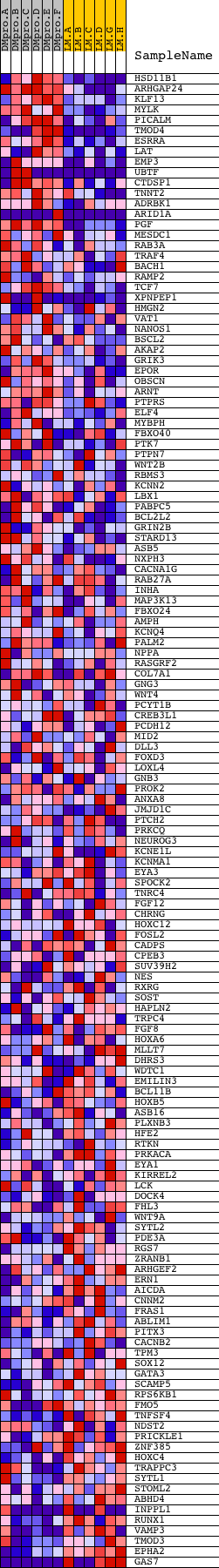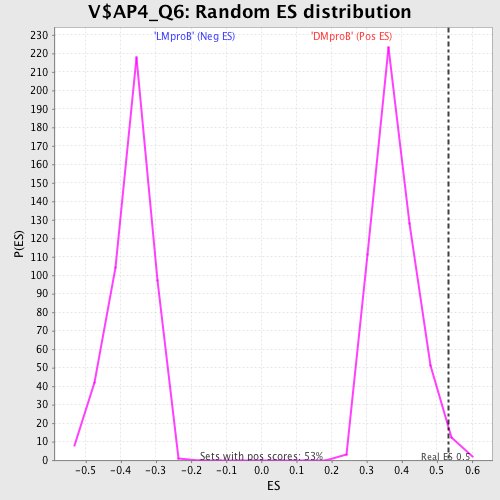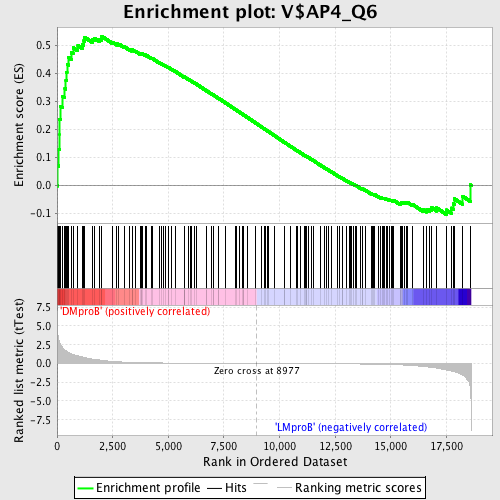
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | DMproB |
| GeneSet | V$AP4_Q6 |
| Enrichment Score (ES) | 0.5333771 |
| Normalized Enrichment Score (NES) | 1.4086614 |
| Nominal p-value | 0.009433962 |
| FDR q-value | 0.4261792 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HSD11B1 | 9125 4019 | 34 | 3.879 | 0.0710 | Yes | ||
| 2 | ARHGAP24 | 16778 3655 | 72 | 3.246 | 0.1299 | Yes | ||
| 3 | KLF13 | 17807 | 103 | 2.916 | 0.1830 | Yes | ||
| 4 | MYLK | 22778 4213 | 107 | 2.894 | 0.2371 | Yes | ||
| 5 | PICALM | 18191 | 149 | 2.592 | 0.2836 | Yes | ||
| 6 | TMOD4 | 1816 1884 15509 | 238 | 2.136 | 0.3189 | Yes | ||
| 7 | ESRRA | 23802 | 350 | 1.777 | 0.3462 | Yes | ||
| 8 | LAT | 17643 | 395 | 1.669 | 0.3752 | Yes | ||
| 9 | EMP3 | 17824 968 | 423 | 1.631 | 0.4043 | Yes | ||
| 10 | UBTF | 1313 10052 | 447 | 1.588 | 0.4329 | Yes | ||
| 11 | CTDSP1 | 4121 10407 | 511 | 1.485 | 0.4573 | Yes | ||
| 12 | TNNT2 | 14113 | 643 | 1.269 | 0.4741 | Yes | ||
| 13 | ADRBK1 | 23960 | 722 | 1.175 | 0.4919 | Yes | ||
| 14 | ARID1A | 8214 2501 13554 | 934 | 1.000 | 0.4993 | Yes | ||
| 15 | PGF | 21020 | 1134 | 0.859 | 0.5046 | Yes | ||
| 16 | MESDC1 | 17771 | 1203 | 0.811 | 0.5162 | Yes | ||
| 17 | RAB3A | 9681 | 1223 | 0.798 | 0.5301 | Yes | ||
| 18 | TRAF4 | 10217 5796 1400 | 1600 | 0.579 | 0.5206 | Yes | ||
| 19 | BACH1 | 8647 | 1684 | 0.542 | 0.5263 | Yes | ||
| 20 | RAMP2 | 20660 | 1923 | 0.448 | 0.5218 | Yes | ||
| 21 | TCF7 | 1467 20466 | 1990 | 0.422 | 0.5262 | Yes | ||
| 22 | XPNPEP1 | 9310 3677 23645 | 2002 | 0.416 | 0.5334 | Yes | ||
| 23 | HMGN2 | 9095 | 2501 | 0.274 | 0.5116 | No | ||
| 24 | VAT1 | 11253 | 2689 | 0.235 | 0.5059 | No | ||
| 25 | NANOS1 | 6858 | 2779 | 0.217 | 0.5051 | No | ||
| 26 | BSCL2 | 23940 | 3015 | 0.185 | 0.4959 | No | ||
| 27 | AKAP2 | 8565 | 3259 | 0.155 | 0.4856 | No | ||
| 28 | GRIK3 | 16083 | 3366 | 0.144 | 0.4826 | No | ||
| 29 | EPOR | 19204 | 3380 | 0.143 | 0.4846 | No | ||
| 30 | OBSCN | 20434 | 3509 | 0.130 | 0.4801 | No | ||
| 31 | ARNT | 4413 1857 | 3754 | 0.112 | 0.4690 | No | ||
| 32 | PTPRS | 1507 1565 1602 22928 | 3776 | 0.111 | 0.4699 | No | ||
| 33 | ELF4 | 24162 | 3803 | 0.108 | 0.4705 | No | ||
| 34 | MYBPH | 14130 | 3850 | 0.105 | 0.4700 | No | ||
| 35 | FBXO40 | 22599 444 | 3951 | 0.099 | 0.4665 | No | ||
| 36 | PTK7 | 22959 | 4032 | 0.094 | 0.4639 | No | ||
| 37 | PTPN7 | 11500 | 4257 | 0.083 | 0.4533 | No | ||
| 38 | WNT2B | 15214 1838 | 4269 | 0.083 | 0.4543 | No | ||
| 39 | RBMS3 | 18973 | 4601 | 0.070 | 0.4377 | No | ||
| 40 | KCNN2 | 8933 1949 | 4670 | 0.068 | 0.4353 | No | ||
| 41 | LBX1 | 98 23659 | 4791 | 0.064 | 0.4300 | No | ||
| 42 | PABPC5 | 24077 | 4873 | 0.062 | 0.4268 | No | ||
| 43 | BCL2L2 | 8653 4441 | 5003 | 0.058 | 0.4209 | No | ||
| 44 | GRIN2B | 16957 | 5009 | 0.058 | 0.4217 | No | ||
| 45 | STARD13 | 10833 | 5150 | 0.055 | 0.4151 | No | ||
| 46 | ASB5 | 13322 | 5310 | 0.051 | 0.4075 | No | ||
| 47 | NXPH3 | 20281 | 5707 | 0.042 | 0.3869 | No | ||
| 48 | CACNA1G | 1237 940 20292 | 5708 | 0.042 | 0.3876 | No | ||
| 49 | RAB27A | 19381 | 5713 | 0.042 | 0.3882 | No | ||
| 50 | INHA | 14214 | 5893 | 0.039 | 0.3793 | No | ||
| 51 | MAP3K13 | 22814 | 5986 | 0.037 | 0.3750 | No | ||
| 52 | FBXO24 | 12918 3649 | 6049 | 0.036 | 0.3723 | No | ||
| 53 | AMPH | 3256 21700 | 6185 | 0.034 | 0.3657 | No | ||
| 54 | KCNQ4 | 12247 2533 | 6252 | 0.033 | 0.3627 | No | ||
| 55 | PALM2 | 10817 2368 | 6694 | 0.026 | 0.3393 | No | ||
| 56 | NPPA | 298 | 6736 | 0.026 | 0.3376 | No | ||
| 57 | RASGRF2 | 21397 | 6954 | 0.023 | 0.3263 | No | ||
| 58 | COL7A1 | 8769 4547 | 7017 | 0.022 | 0.3233 | No | ||
| 59 | GNG3 | 23752 | 7024 | 0.022 | 0.3234 | No | ||
| 60 | WNT4 | 16025 | 7242 | 0.019 | 0.3120 | No | ||
| 61 | PCYT1B | 24289 | 7275 | 0.019 | 0.3106 | No | ||
| 62 | CREB3L1 | 14521 | 7546 | 0.016 | 0.2963 | No | ||
| 63 | PCDH12 | 7005 | 8001 | 0.011 | 0.2719 | No | ||
| 64 | MID2 | 24232 | 8077 | 0.010 | 0.2681 | No | ||
| 65 | DLL3 | 17912 | 8213 | 0.008 | 0.2609 | No | ||
| 66 | FOXD3 | 9088 | 8324 | 0.007 | 0.2551 | No | ||
| 67 | LOXL4 | 23672 | 8392 | 0.006 | 0.2516 | No | ||
| 68 | GNB3 | 9027 | 8395 | 0.006 | 0.2516 | No | ||
| 69 | PROK2 | 17057 | 8556 | 0.004 | 0.2430 | No | ||
| 70 | ANXA8 | 22046 | 8927 | 0.001 | 0.2230 | No | ||
| 71 | JMJD1C | 11531 19996 | 9199 | -0.002 | 0.2083 | No | ||
| 72 | PTCH2 | 9640 | 9338 | -0.004 | 0.2009 | No | ||
| 73 | PRKCQ | 2873 2831 | 9370 | -0.004 | 0.1993 | No | ||
| 74 | NEUROG3 | 20005 | 9459 | -0.005 | 0.1947 | No | ||
| 75 | KCNE1L | 24044 | 9501 | -0.005 | 0.1925 | No | ||
| 76 | KCNMA1 | 4947 | 9754 | -0.008 | 0.1790 | No | ||
| 77 | EYA3 | 8936 2352 | 10210 | -0.012 | 0.1546 | No | ||
| 78 | SPOCK2 | 13585 | 10217 | -0.012 | 0.1545 | No | ||
| 79 | TNRC4 | 15514 | 10503 | -0.015 | 0.1394 | No | ||
| 80 | FGF12 | 1723 22621 | 10779 | -0.018 | 0.1249 | No | ||
| 81 | CHRNG | 4323 | 10794 | -0.019 | 0.1245 | No | ||
| 82 | HOXC12 | 22343 | 10921 | -0.020 | 0.1180 | No | ||
| 83 | FOSL2 | 4733 8978 16878 | 11141 | -0.023 | 0.1066 | No | ||
| 84 | CADPS | 11298 | 11142 | -0.023 | 0.1070 | No | ||
| 85 | CPEB3 | 5539 | 11190 | -0.023 | 0.1049 | No | ||
| 86 | SUV39H2 | 7232 12263 | 11228 | -0.024 | 0.1033 | No | ||
| 87 | NES | 5162 9454 | 11283 | -0.024 | 0.1009 | No | ||
| 88 | RXRG | 14063 | 11448 | -0.026 | 0.0925 | No | ||
| 89 | SOST | 20210 | 11516 | -0.027 | 0.0894 | No | ||
| 90 | HAPLN2 | 15296 | 11821 | -0.031 | 0.0735 | No | ||
| 91 | TRPC4 | 15593 | 12023 | -0.034 | 0.0633 | No | ||
| 92 | FGF8 | 23656 | 12104 | -0.035 | 0.0596 | No | ||
| 93 | HOXA6 | 17150 | 12196 | -0.037 | 0.0554 | No | ||
| 94 | MLLT7 | 24278 | 12337 | -0.039 | 0.0485 | No | ||
| 95 | DHRS3 | 15997 | 12599 | -0.043 | 0.0352 | No | ||
| 96 | WDTC1 | 10512 | 12696 | -0.045 | 0.0309 | No | ||
| 97 | EMILIN3 | 11376 | 12814 | -0.047 | 0.0254 | No | ||
| 98 | BCL11B | 7190 | 12823 | -0.047 | 0.0259 | No | ||
| 99 | HOXB5 | 20687 | 12998 | -0.051 | 0.0174 | No | ||
| 100 | ASB16 | 20644 | 13146 | -0.054 | 0.0105 | No | ||
| 101 | PLXNB3 | 24304 | 13202 | -0.055 | 0.0085 | No | ||
| 102 | HFE2 | 1777 15495 | 13233 | -0.056 | 0.0080 | No | ||
| 103 | RTKN | 17394 1156 | 13333 | -0.059 | 0.0037 | No | ||
| 104 | PRKACA | 18549 3844 | 13424 | -0.061 | -0.0000 | No | ||
| 105 | EYA1 | 4695 4061 | 13446 | -0.062 | -0.0000 | No | ||
| 106 | KIRREL2 | 17889 3922 | 13634 | -0.068 | -0.0089 | No | ||
| 107 | LCK | 15746 | 13709 | -0.070 | -0.0115 | No | ||
| 108 | DOCK4 | 10705 | 13711 | -0.070 | -0.0103 | No | ||
| 109 | FHL3 | 16092 2457 | 13876 | -0.077 | -0.0177 | No | ||
| 110 | WNT9A | 5709 | 14127 | -0.086 | -0.0296 | No | ||
| 111 | SYTL2 | 18190 1539 3730 | 14183 | -0.088 | -0.0309 | No | ||
| 112 | PDE3A | 7045 | 14207 | -0.089 | -0.0305 | No | ||
| 113 | RGS7 | 13743 | 14285 | -0.093 | -0.0329 | No | ||
| 114 | ZRANB1 | 6884 11702 | 14442 | -0.100 | -0.0395 | No | ||
| 115 | ARHGEF2 | 4987 | 14545 | -0.106 | -0.0430 | No | ||
| 116 | ERN1 | 8137 | 14637 | -0.111 | -0.0459 | No | ||
| 117 | AICDA | 17295 1087 | 14659 | -0.112 | -0.0449 | No | ||
| 118 | CNNM2 | 8251 | 14705 | -0.114 | -0.0452 | No | ||
| 119 | FRAS1 | 16788 3589 | 14799 | -0.120 | -0.0480 | No | ||
| 120 | ABLIM1 | 3717 3766 3679 | 14852 | -0.124 | -0.0485 | No | ||
| 121 | PITX3 | 9571 | 14953 | -0.132 | -0.0514 | No | ||
| 122 | CACNB2 | 4466 8678 | 15022 | -0.138 | -0.0525 | No | ||
| 123 | TPM3 | 12233 7209 7208 1790 | 15087 | -0.143 | -0.0533 | No | ||
| 124 | SOX12 | 5478 | 15134 | -0.147 | -0.0530 | No | ||
| 125 | GATA3 | 9004 | 15431 | -0.179 | -0.0657 | No | ||
| 126 | SCAMP5 | 3028 19100 | 15438 | -0.180 | -0.0626 | No | ||
| 127 | RPS6KB1 | 7815 1207 13040 | 15488 | -0.186 | -0.0618 | No | ||
| 128 | FMO5 | 4730 15489 | 15527 | -0.193 | -0.0602 | No | ||
| 129 | TNFSF4 | 14082 | 15621 | -0.206 | -0.0614 | No | ||
| 130 | NDST2 | 21906 | 15694 | -0.216 | -0.0612 | No | ||
| 131 | PRICKLE1 | 8379 | 15752 | -0.225 | -0.0601 | No | ||
| 132 | ZNF385 | 22106 2297 | 15980 | -0.267 | -0.0674 | No | ||
| 133 | HOXC4 | 4864 | 16454 | -0.386 | -0.0857 | No | ||
| 134 | TRAPPC3 | 6553 16080 2469 11305 | 16621 | -0.435 | -0.0865 | No | ||
| 135 | SYTL1 | 15728 | 16739 | -0.483 | -0.0838 | No | ||
| 136 | STOML2 | 15902 | 16834 | -0.529 | -0.0790 | No | ||
| 137 | ABHD4 | 22021 | 17063 | -0.618 | -0.0797 | No | ||
| 138 | INPPL1 | 17733 3775 | 17500 | -0.863 | -0.0871 | No | ||
| 139 | RUNX1 | 4481 | 17719 | -1.000 | -0.0801 | No | ||
| 140 | VAMP3 | 15660 | 17811 | -1.054 | -0.0653 | No | ||
| 141 | TMOD3 | 6945 | 17860 | -1.088 | -0.0474 | No | ||
| 142 | EPHA2 | 16006 | 18224 | -1.564 | -0.0377 | No | ||
| 143 | GAS7 | 20836 1299 | 18571 | -3.138 | 0.0024 | No |