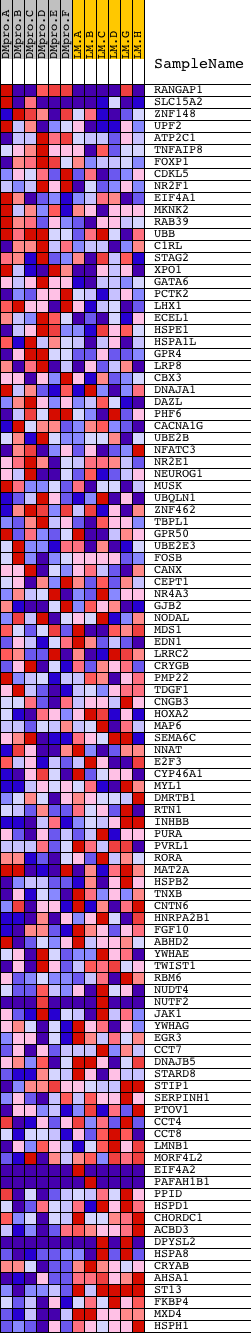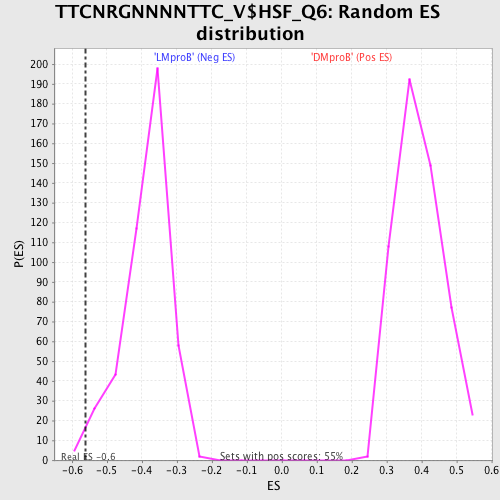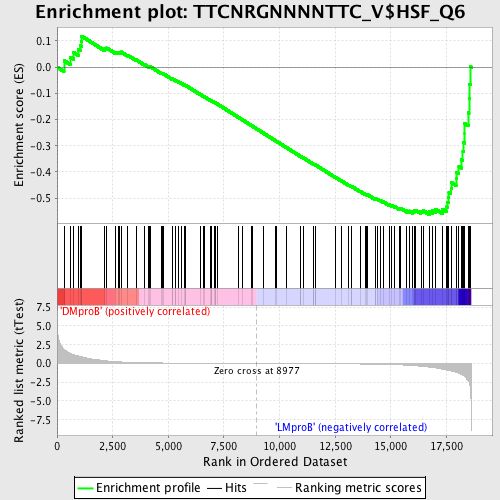
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | TTCNRGNNNNTTC_V$HSF_Q6 |
| Enrichment Score (ES) | -0.5624742 |
| Normalized Enrichment Score (NES) | -1.4553852 |
| Nominal p-value | 0.011135858 |
| FDR q-value | 0.572503 |
| FWER p-Value | 0.974 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RANGAP1 | 2180 22195 | 310 | 1.906 | 0.0252 | No | ||
| 2 | SLC15A2 | 22600 | 613 | 1.309 | 0.0377 | No | ||
| 3 | ZNF148 | 5949 | 749 | 1.147 | 0.0557 | No | ||
| 4 | UPF2 | 11591 | 953 | 0.994 | 0.0666 | No | ||
| 5 | ATP2C1 | 10660 6178 | 1034 | 0.935 | 0.0828 | No | ||
| 6 | TNFAIP8 | 8391 | 1089 | 0.898 | 0.0997 | No | ||
| 7 | FOXP1 | 4242 | 1101 | 0.886 | 0.1186 | No | ||
| 8 | CDKL5 | 24019 | 2146 | 0.368 | 0.0703 | No | ||
| 9 | NR2F1 | 7015 21401 | 2232 | 0.345 | 0.0733 | No | ||
| 10 | EIF4A1 | 8889 23719 | 2637 | 0.245 | 0.0569 | No | ||
| 11 | MKNK2 | 3299 9392 | 2749 | 0.222 | 0.0558 | No | ||
| 12 | RAB39 | 19113 | 2822 | 0.210 | 0.0565 | No | ||
| 13 | UBB | 10240 | 2874 | 0.202 | 0.0582 | No | ||
| 14 | C1RL | 10559 17288 | 3167 | 0.165 | 0.0460 | No | ||
| 15 | STAG2 | 5521 | 3550 | 0.126 | 0.0282 | No | ||
| 16 | XPO1 | 4172 | 3925 | 0.101 | 0.0102 | No | ||
| 17 | GATA6 | 96 | 4108 | 0.090 | 0.0024 | No | ||
| 18 | PCTK2 | 19907 | 4135 | 0.089 | 0.0029 | No | ||
| 19 | LHX1 | 4995 | 4180 | 0.087 | 0.0025 | No | ||
| 20 | ECEL1 | 13893 | 4700 | 0.067 | -0.0241 | No | ||
| 21 | HSPE1 | 9133 | 4717 | 0.067 | -0.0235 | No | ||
| 22 | HSPA1L | 23267 | 4803 | 0.063 | -0.0267 | No | ||
| 23 | GPR4 | 18364 | 5170 | 0.054 | -0.0453 | No | ||
| 24 | LRP8 | 16149 2425 | 5184 | 0.054 | -0.0448 | No | ||
| 25 | CBX3 | 8705 4484 | 5334 | 0.050 | -0.0517 | No | ||
| 26 | DNAJA1 | 4878 16242 2345 | 5340 | 0.050 | -0.0509 | No | ||
| 27 | DAZL | 22944 | 5463 | 0.047 | -0.0565 | No | ||
| 28 | PHF6 | 2648 24340 | 5571 | 0.045 | -0.0612 | No | ||
| 29 | CACNA1G | 1237 940 20292 | 5708 | 0.042 | -0.0677 | No | ||
| 30 | UBE2B | 10245 | 5750 | 0.042 | -0.0689 | No | ||
| 31 | NFATC3 | 5169 | 5784 | 0.041 | -0.0698 | No | ||
| 32 | NR2E1 | 19780 | 6444 | 0.030 | -0.1048 | No | ||
| 33 | NEUROG1 | 21446 | 6576 | 0.028 | -0.1112 | No | ||
| 34 | MUSK | 5199 | 6603 | 0.027 | -0.1120 | No | ||
| 35 | UBQLN1 | 3237 3209 3248 3291 | 6911 | 0.023 | -0.1281 | No | ||
| 36 | ZNF462 | 6321 | 6925 | 0.023 | -0.1283 | No | ||
| 37 | TBPL1 | 19805 | 6935 | 0.023 | -0.1283 | No | ||
| 38 | GPR50 | 24318 | 7068 | 0.021 | -0.1349 | No | ||
| 39 | UBE2E3 | 5816 5815 | 7109 | 0.021 | -0.1366 | No | ||
| 40 | FOSB | 17945 | 7128 | 0.021 | -0.1372 | No | ||
| 41 | CANX | 4474 | 7196 | 0.020 | -0.1403 | No | ||
| 42 | CEPT1 | 13624 | 8167 | 0.009 | -0.1926 | No | ||
| 43 | NR4A3 | 9473 16212 5183 | 8349 | 0.006 | -0.2022 | No | ||
| 44 | GJB2 | 9018 21806 | 8735 | 0.002 | -0.2229 | No | ||
| 45 | NODAL | 333 20010 | 8777 | 0.002 | -0.2251 | No | ||
| 46 | MDS1 | 9375 | 9267 | -0.003 | -0.2514 | No | ||
| 47 | EDN1 | 21658 | 9812 | -0.008 | -0.2807 | No | ||
| 48 | LRRC2 | 13189 | 9879 | -0.009 | -0.2840 | No | ||
| 49 | CRYGB | 8795 | 10326 | -0.013 | -0.3078 | No | ||
| 50 | PMP22 | 9594 1312 | 10924 | -0.020 | -0.3396 | No | ||
| 51 | TDGF1 | 5704 10108 | 10947 | -0.020 | -0.3404 | No | ||
| 52 | CNGB3 | 16267 | 11056 | -0.022 | -0.3457 | No | ||
| 53 | HOXA2 | 17151 | 11511 | -0.027 | -0.3697 | No | ||
| 54 | MAP6 | 9419 | 11544 | -0.028 | -0.3708 | No | ||
| 55 | SEMA6C | 1797 5424 | 11595 | -0.028 | -0.3729 | No | ||
| 56 | NNAT | 2764 5180 | 11612 | -0.029 | -0.3731 | No | ||
| 57 | E2F3 | 21500 8874 | 12512 | -0.042 | -0.4207 | No | ||
| 58 | CYP46A1 | 21158 | 12789 | -0.047 | -0.4346 | No | ||
| 59 | MYL1 | 4015 | 13102 | -0.053 | -0.4503 | No | ||
| 60 | DMRTB1 | 15813 | 13214 | -0.056 | -0.4551 | No | ||
| 61 | RTN1 | 4177 2099 | 13253 | -0.057 | -0.4559 | No | ||
| 62 | INHBB | 13858 | 13629 | -0.068 | -0.4746 | No | ||
| 63 | PURA | 9670 | 13841 | -0.076 | -0.4844 | No | ||
| 64 | PVRL1 | 3006 19482 12211 7192 | 13915 | -0.078 | -0.4866 | No | ||
| 65 | RORA | 3019 5388 | 13938 | -0.079 | -0.4860 | No | ||
| 66 | MAT2A | 10553 10554 6099 | 14291 | -0.093 | -0.5030 | No | ||
| 67 | HSPB2 | 19121 | 14294 | -0.093 | -0.5011 | No | ||
| 68 | TNXB | 1557 8174 878 8175 | 14382 | -0.097 | -0.5036 | No | ||
| 69 | CNTN6 | 17346 | 14521 | -0.105 | -0.5088 | No | ||
| 70 | HNRPA2B1 | 1187 7001 7000 1040 | 14665 | -0.112 | -0.5140 | No | ||
| 71 | FGF10 | 4719 8962 | 14928 | -0.130 | -0.5253 | No | ||
| 72 | ABHD2 | 18208 12043 | 15030 | -0.138 | -0.5277 | No | ||
| 73 | YWHAE | 20776 | 15156 | -0.149 | -0.5312 | No | ||
| 74 | TWIST1 | 21291 | 15409 | -0.176 | -0.5409 | No | ||
| 75 | RBM6 | 9707 3095 3063 2995 | 15453 | -0.182 | -0.5393 | No | ||
| 76 | NUDT4 | 19639 | 15713 | -0.218 | -0.5484 | No | ||
| 77 | NUTF2 | 12632 7529 | 15824 | -0.238 | -0.5491 | No | ||
| 78 | JAK1 | 15827 | 15986 | -0.269 | -0.5519 | No | ||
| 79 | YWHAG | 16339 | 16043 | -0.282 | -0.5488 | No | ||
| 80 | EGR3 | 4656 | 16111 | -0.298 | -0.5458 | No | ||
| 81 | CCT7 | 17385 | 16356 | -0.361 | -0.5511 | No | ||
| 82 | DNAJB5 | 16236 | 16448 | -0.385 | -0.5475 | No | ||
| 83 | STARD8 | 10682 | 16726 | -0.475 | -0.5520 | Yes | ||
| 84 | STIP1 | 23798 | 16884 | -0.547 | -0.5484 | Yes | ||
| 85 | SERPINH1 | 8703 | 17002 | -0.588 | -0.5418 | Yes | ||
| 86 | PTOV1 | 13522 | 17326 | -0.756 | -0.5426 | Yes | ||
| 87 | CCT4 | 8710 | 17518 | -0.872 | -0.5338 | Yes | ||
| 88 | CCT8 | 22551 | 17532 | -0.882 | -0.5151 | Yes | ||
| 89 | LMNB1 | 23549 1943 | 17584 | -0.910 | -0.4978 | Yes | ||
| 90 | MORF4L2 | 12118 | 17612 | -0.932 | -0.4787 | Yes | ||
| 91 | EIF4A2 | 4660 1679 1645 | 17704 | -0.997 | -0.4617 | Yes | ||
| 92 | PAFAH1B1 | 1340 5220 9524 | 17724 | -1.001 | -0.4407 | Yes | ||
| 93 | PPID | 12576 | 17933 | -1.153 | -0.4266 | Yes | ||
| 94 | HSPD1 | 4078 9129 | 17940 | -1.159 | -0.4014 | Yes | ||
| 95 | CHORDC1 | 12430 | 18061 | -1.286 | -0.3795 | Yes | ||
| 96 | ACBD3 | 14025 | 18170 | -1.461 | -0.3532 | Yes | ||
| 97 | DPYSL2 | 3122 4561 8787 | 18242 | -1.581 | -0.3222 | Yes | ||
| 98 | HSPA8 | 9124 3072 4871 | 18273 | -1.625 | -0.2881 | Yes | ||
| 99 | CRYAB | 19457 | 18316 | -1.702 | -0.2529 | Yes | ||
| 100 | AHSA1 | 21192 | 18318 | -1.710 | -0.2153 | Yes | ||
| 101 | ST13 | 12865 | 18484 | -2.298 | -0.1737 | Yes | ||
| 102 | FKBP4 | 4726 | 18514 | -2.486 | -0.1205 | Yes | ||
| 103 | MXD4 | 9346 | 18525 | -2.578 | -0.0643 | Yes | ||
| 104 | HSPH1 | 16282 | 18572 | -3.143 | 0.0024 | Yes |