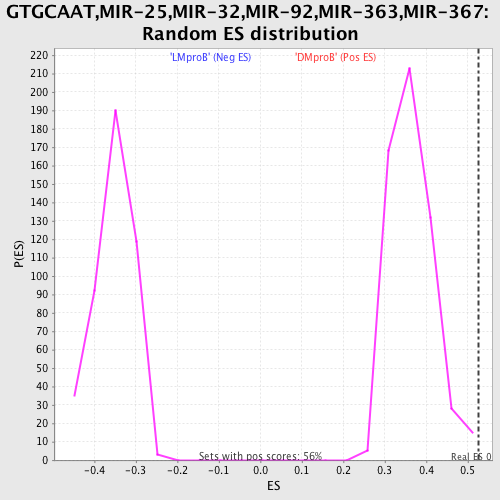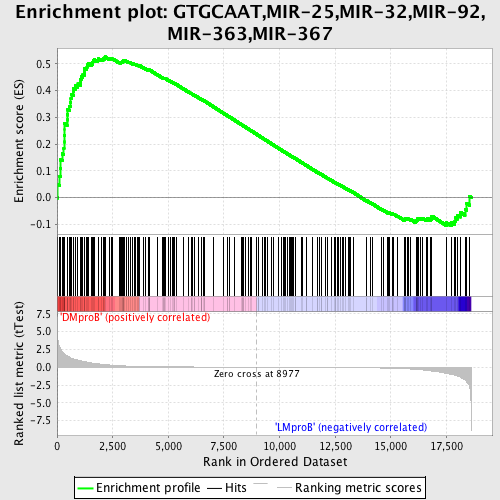
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | DMproB |
| GeneSet | GTGCAAT,MIR-25,MIR-32,MIR-92,MIR-363,MIR-367 |
| Enrichment Score (ES) | 0.5263717 |
| Normalized Enrichment Score (NES) | 1.4466631 |
| Nominal p-value | 0.0053475937 |
| FDR q-value | 0.4296772 |
| FWER p-Value | 0.989 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TRAF3 | 21147 | 30 | 4.000 | 0.0485 | Yes | ||
| 2 | ITPR1 | 17341 | 120 | 2.791 | 0.0786 | Yes | ||
| 3 | BCL11A | 4691 | 147 | 2.605 | 0.1098 | Yes | ||
| 4 | FNBP4 | 14956 2671 | 159 | 2.550 | 0.1411 | Yes | ||
| 5 | OGT | 4241 24274 | 234 | 2.156 | 0.1641 | Yes | ||
| 6 | PIK3AP1 | 23679 | 303 | 1.929 | 0.1846 | Yes | ||
| 7 | ATP2A2 | 4421 3481 | 311 | 1.906 | 0.2081 | Yes | ||
| 8 | CDKN1C | 17546 | 315 | 1.898 | 0.2317 | Yes | ||
| 9 | PITPNA | 20778 | 329 | 1.853 | 0.2542 | Yes | ||
| 10 | BCAT2 | 18244 | 331 | 1.841 | 0.2772 | Yes | ||
| 11 | FBXW7 | 1805 11928 | 450 | 1.581 | 0.2906 | Yes | ||
| 12 | PIP5K1C | 5250 19929 | 458 | 1.572 | 0.3099 | Yes | ||
| 13 | REV3L | 20050 | 478 | 1.541 | 0.3281 | Yes | ||
| 14 | TOB2 | 7162 | 551 | 1.417 | 0.3420 | Yes | ||
| 15 | POLS | 9963 | 596 | 1.338 | 0.3563 | Yes | ||
| 16 | UGP2 | 20518 | 621 | 1.295 | 0.3712 | Yes | ||
| 17 | MCL1 | 15502 | 654 | 1.254 | 0.3852 | Yes | ||
| 18 | CIC | 18338 | 733 | 1.162 | 0.3955 | Yes | ||
| 19 | FMN2 | 14037 | 742 | 1.155 | 0.4096 | Yes | ||
| 20 | ADM | 18125 | 815 | 1.086 | 0.4192 | Yes | ||
| 21 | SLC38A2 | 22149 2219 | 937 | 1.000 | 0.4252 | Yes | ||
| 22 | RPS6KA4 | 23804 | 1047 | 0.927 | 0.4309 | Yes | ||
| 23 | BAZ2A | 4371 | 1059 | 0.921 | 0.4418 | Yes | ||
| 24 | ARRDC4 | 12346 | 1083 | 0.907 | 0.4519 | Yes | ||
| 25 | MYO18A | 20766 | 1155 | 0.845 | 0.4587 | Yes | ||
| 26 | BTBD12 | 22684 | 1217 | 0.800 | 0.4654 | Yes | ||
| 27 | SOX4 | 5479 | 1229 | 0.790 | 0.4746 | Yes | ||
| 28 | DAB2IP | 12808 2692 | 1237 | 0.786 | 0.4841 | Yes | ||
| 29 | MAN2A1 | 23170 | 1339 | 0.721 | 0.4877 | Yes | ||
| 30 | RNF44 | 8364 | 1347 | 0.717 | 0.4963 | Yes | ||
| 31 | PPP1R12A | 3354 19886 | 1421 | 0.672 | 0.5007 | Yes | ||
| 32 | TOB1 | 20703 | 1542 | 0.608 | 0.5018 | Yes | ||
| 33 | RAP1B | 10083 | 1596 | 0.582 | 0.5062 | Yes | ||
| 34 | SRPR | 12525 7433 | 1613 | 0.573 | 0.5125 | Yes | ||
| 35 | VPS54 | 20941 | 1663 | 0.553 | 0.5168 | Yes | ||
| 36 | RAB14 | 12685 | 1837 | 0.483 | 0.5134 | Yes | ||
| 37 | PCTK1 | 24369 | 1847 | 0.479 | 0.5189 | Yes | ||
| 38 | HERC2 | 9085 | 1988 | 0.423 | 0.5166 | Yes | ||
| 39 | LHFPL2 | 21584 | 2064 | 0.392 | 0.5175 | Yes | ||
| 40 | P2RY13 | 15332 | 2094 | 0.383 | 0.5207 | Yes | ||
| 41 | ZDHHC5 | 14547 | 2147 | 0.367 | 0.5225 | Yes | ||
| 42 | HIVEP1 | 8486 | 2160 | 0.364 | 0.5264 | Yes | ||
| 43 | MAP2K4 | 20405 | 2372 | 0.306 | 0.5187 | No | ||
| 44 | PHTF2 | 16599 | 2443 | 0.288 | 0.5186 | No | ||
| 45 | SLC32A1 | 14757 | 2506 | 0.274 | 0.5186 | No | ||
| 46 | ITGA5 | 22105 | 2790 | 0.214 | 0.5059 | No | ||
| 47 | KLF4 | 9230 4962 4963 | 2856 | 0.205 | 0.5050 | No | ||
| 48 | TEF | 2287 22414 2192 | 2875 | 0.202 | 0.5065 | No | ||
| 49 | BRMS1L | 21268 | 2916 | 0.197 | 0.5068 | No | ||
| 50 | KLF2 | 9229 | 2924 | 0.196 | 0.5089 | No | ||
| 51 | PTPRO | 1093 17256 | 2962 | 0.192 | 0.5093 | No | ||
| 52 | NFIX | 9458 | 2978 | 0.190 | 0.5109 | No | ||
| 53 | FBN1 | 14449 | 2987 | 0.189 | 0.5128 | No | ||
| 54 | TSGA14 | 17194 | 3030 | 0.183 | 0.5128 | No | ||
| 55 | STK39 | 14570 | 3108 | 0.174 | 0.5108 | No | ||
| 56 | DDX3X | 24379 | 3230 | 0.158 | 0.5062 | No | ||
| 57 | NFIA | 16172 5170 | 3280 | 0.153 | 0.5054 | No | ||
| 58 | DLGAP2 | 10856 6350 | 3399 | 0.141 | 0.5008 | No | ||
| 59 | ADAM19 | 8548 | 3458 | 0.136 | 0.4993 | No | ||
| 60 | MYLIP | 21652 3189 | 3504 | 0.131 | 0.4985 | No | ||
| 61 | MYH9 | 2252 2244 | 3601 | 0.122 | 0.4948 | No | ||
| 62 | SLC24A3 | 14818 | 3649 | 0.119 | 0.4938 | No | ||
| 63 | TCF2 | 1395 20727 | 3688 | 0.116 | 0.4932 | No | ||
| 64 | LYST | 9325 5040 | 3701 | 0.115 | 0.4940 | No | ||
| 65 | KLHL18 | 11291 | 3865 | 0.104 | 0.4864 | No | ||
| 66 | ARMC1 | 7907 | 3973 | 0.098 | 0.4818 | No | ||
| 67 | GATA6 | 96 | 4108 | 0.090 | 0.4757 | No | ||
| 68 | CREB1 | 3990 8782 4558 4093 | 4110 | 0.090 | 0.4767 | No | ||
| 69 | IXL | 17910 | 4133 | 0.089 | 0.4767 | No | ||
| 70 | DDX3Y | 6507 4593 | 4142 | 0.089 | 0.4773 | No | ||
| 71 | SDC2 | 9134 | 4161 | 0.088 | 0.4775 | No | ||
| 72 | SSFA2 | 7692 | 4506 | 0.074 | 0.4597 | No | ||
| 73 | PITPNC1 | 7759 | 4718 | 0.066 | 0.4491 | No | ||
| 74 | LBX1 | 98 23659 | 4791 | 0.064 | 0.4460 | No | ||
| 75 | BMPR2 | 14241 4081 | 4800 | 0.063 | 0.4463 | No | ||
| 76 | TACC2 | 18052 2534 2234 2435 2350 | 4809 | 0.063 | 0.4467 | No | ||
| 77 | IBTK | 19047 | 4817 | 0.063 | 0.4471 | No | ||
| 78 | SMAD7 | 23513 | 4874 | 0.062 | 0.4448 | No | ||
| 79 | RHPN2 | 18293 | 5022 | 0.058 | 0.4376 | No | ||
| 80 | FHL2 | 13966 | 5091 | 0.056 | 0.4346 | No | ||
| 81 | SCN3A | 14573 | 5175 | 0.054 | 0.4307 | No | ||
| 82 | CNTN4 | 11265 1088 17345 | 5253 | 0.052 | 0.4272 | No | ||
| 83 | PER2 | 3940 13881 | 5265 | 0.052 | 0.4273 | No | ||
| 84 | CEBPA | 4514 | 5270 | 0.052 | 0.4277 | No | ||
| 85 | LUZP1 | 6533 11257 2503 | 5372 | 0.049 | 0.4228 | No | ||
| 86 | SMAD6 | 19083 | 5682 | 0.043 | 0.4066 | No | ||
| 87 | COL1A2 | 8772 | 5895 | 0.039 | 0.3956 | No | ||
| 88 | RAD21 | 22298 | 6019 | 0.037 | 0.3893 | No | ||
| 89 | NPTX1 | 20126 | 6039 | 0.037 | 0.3888 | No | ||
| 90 | PAX3 | 13910 | 6082 | 0.036 | 0.3869 | No | ||
| 91 | RNF141 | 12478 7392 | 6160 | 0.034 | 0.3832 | No | ||
| 92 | DNAJB9 | 6565 | 6184 | 0.034 | 0.3824 | No | ||
| 93 | EDEM1 | 17339 938 | 6349 | 0.031 | 0.3738 | No | ||
| 94 | PRKCE | 9575 | 6468 | 0.029 | 0.3678 | No | ||
| 95 | PRDM13 | 15933 | 6494 | 0.029 | 0.3668 | No | ||
| 96 | COL12A1 | 3143 19054 | 6499 | 0.029 | 0.3669 | No | ||
| 97 | TRAM2 | 13990 | 6597 | 0.027 | 0.3620 | No | ||
| 98 | RSBN1 | 6035 | 6614 | 0.027 | 0.3615 | No | ||
| 99 | NFIB | 15855 | 6619 | 0.027 | 0.3616 | No | ||
| 100 | GOLGA4 | 12022 | 7025 | 0.022 | 0.3399 | No | ||
| 101 | ITGAV | 4931 | 7467 | 0.017 | 0.3161 | No | ||
| 102 | BTG2 | 13832 | 7652 | 0.015 | 0.3063 | No | ||
| 103 | TEAD1 | 18121 | 7667 | 0.014 | 0.3057 | No | ||
| 104 | RNF38 | 7862 2328 13115 | 7731 | 0.014 | 0.3025 | No | ||
| 105 | PLEKHA6 | 10786 6288 | 7769 | 0.013 | 0.3006 | No | ||
| 106 | HAS3 | 18477 | 7983 | 0.011 | 0.2892 | No | ||
| 107 | ATXN3 | 21001 2051 | 8290 | 0.007 | 0.2727 | No | ||
| 108 | NR4A3 | 9473 16212 5183 | 8349 | 0.006 | 0.2696 | No | ||
| 109 | ROBO2 | 6501 | 8351 | 0.006 | 0.2696 | No | ||
| 110 | TCF21 | 3437 | 8374 | 0.006 | 0.2685 | No | ||
| 111 | ADAM10 | 4336 | 8389 | 0.006 | 0.2678 | No | ||
| 112 | DPP10 | 13850 | 8457 | 0.005 | 0.2643 | No | ||
| 113 | CXCL5 | 16802 | 8582 | 0.004 | 0.2576 | No | ||
| 114 | FMR1 | 24321 | 8683 | 0.003 | 0.2522 | No | ||
| 115 | CD2AP | 22975 | 8739 | 0.002 | 0.2492 | No | ||
| 116 | CXXC5 | 12523 | 8941 | 0.000 | 0.2383 | No | ||
| 117 | ELOVL4 | 19048 | 8943 | 0.000 | 0.2383 | No | ||
| 118 | ADCY3 | 21329 894 | 9063 | -0.001 | 0.2318 | No | ||
| 119 | SIM2 | 22693 | 9072 | -0.001 | 0.2314 | No | ||
| 120 | RGS17 | 7133 | 9214 | -0.003 | 0.2238 | No | ||
| 121 | SOX11 | 5477 | 9302 | -0.004 | 0.2191 | No | ||
| 122 | USP28 | 19465 | 9371 | -0.004 | 0.2154 | No | ||
| 123 | EXOC5 | 8370 | 9466 | -0.005 | 0.2104 | No | ||
| 124 | SLC12A5 | 14731 | 9641 | -0.007 | 0.2010 | No | ||
| 125 | MARK1 | 5955 | 9717 | -0.007 | 0.1970 | No | ||
| 126 | EDG1 | 15187 | 9969 | -0.010 | 0.1835 | No | ||
| 127 | KIF5B | 4955 2031 | 10080 | -0.011 | 0.1777 | No | ||
| 128 | HOXB8 | 9111 | 10175 | -0.012 | 0.1727 | No | ||
| 129 | CNNM4 | 14273 | 10239 | -0.012 | 0.1695 | No | ||
| 130 | CSMD3 | 10735 7938 | 10276 | -0.013 | 0.1677 | No | ||
| 131 | HAND1 | 20440 | 10349 | -0.013 | 0.1639 | No | ||
| 132 | ARHGEF17 | 17737 | 10435 | -0.014 | 0.1595 | No | ||
| 133 | CREB3L2 | 17182 | 10494 | -0.015 | 0.1565 | No | ||
| 134 | SOCS5 | 1619 23141 | 10533 | -0.015 | 0.1546 | No | ||
| 135 | EGR2 | 8886 | 10558 | -0.016 | 0.1535 | No | ||
| 136 | XYLT2 | 20288 | 10616 | -0.016 | 0.1506 | No | ||
| 137 | DSC2 | 8865 | 10706 | -0.018 | 0.1460 | No | ||
| 138 | OAZ3 | 15255 | 10717 | -0.018 | 0.1457 | No | ||
| 139 | PCDH11X | 2559 | 10963 | -0.020 | 0.1327 | No | ||
| 140 | CBLN4 | 14326 | 11035 | -0.021 | 0.1291 | No | ||
| 141 | CPEB3 | 5539 | 11190 | -0.023 | 0.1210 | No | ||
| 142 | RFX1 | 18548 | 11218 | -0.024 | 0.1198 | No | ||
| 143 | TRAK2 | 12903 13946 | 11475 | -0.027 | 0.1063 | No | ||
| 144 | WRNIP1 | 21675 3161 | 11690 | -0.030 | 0.0950 | No | ||
| 145 | ATRX | 10350 5922 2628 | 11722 | -0.030 | 0.0937 | No | ||
| 146 | HHIP | 4849 | 11781 | -0.031 | 0.0909 | No | ||
| 147 | RBPMS2 | 19399 | 11892 | -0.032 | 0.0854 | No | ||
| 148 | GFPT2 | 4775 | 11903 | -0.033 | 0.0852 | No | ||
| 149 | SERTAD2 | 12200 7182 | 12059 | -0.035 | 0.0772 | No | ||
| 150 | MOAP1 | 12252 | 12153 | -0.036 | 0.0726 | No | ||
| 151 | FZD10 | 13565 16693 | 12160 | -0.036 | 0.0728 | No | ||
| 152 | MYO1B | 13962 | 12347 | -0.039 | 0.0632 | No | ||
| 153 | DUSP10 | 4003 14016 | 12469 | -0.041 | 0.0571 | No | ||
| 154 | E2F3 | 21500 8874 | 12512 | -0.042 | 0.0553 | No | ||
| 155 | NOX4 | 18196 | 12532 | -0.042 | 0.0548 | No | ||
| 156 | HAND2 | 18615 | 12607 | -0.043 | 0.0514 | No | ||
| 157 | SLC17A6 | 18226 | 12629 | -0.044 | 0.0508 | No | ||
| 158 | GAP43 | 9001 | 12751 | -0.046 | 0.0448 | No | ||
| 159 | SYN2 | 5559 | 12822 | -0.047 | 0.0416 | No | ||
| 160 | CCNC | 16263 | 12879 | -0.048 | 0.0391 | No | ||
| 161 | RAB23 | 14282 | 12962 | -0.050 | 0.0353 | No | ||
| 162 | PTGER4 | 9649 | 13093 | -0.053 | 0.0289 | No | ||
| 163 | WBSCR18 | 7254 12289 | 13145 | -0.054 | 0.0268 | No | ||
| 164 | LATS2 | 11923 6928 | 13206 | -0.056 | 0.0243 | No | ||
| 165 | EPHA8 | 15705 | 13316 | -0.058 | 0.0191 | No | ||
| 166 | BSN | 18997 | 13895 | -0.078 | -0.0114 | No | ||
| 167 | GDF11 | 9011 | 13927 | -0.079 | -0.0121 | No | ||
| 168 | CPEB2 | 6077 | 14064 | -0.084 | -0.0184 | No | ||
| 169 | SNF1LK | 23033 | 14166 | -0.088 | -0.0228 | No | ||
| 170 | DSCAML1 | 4338 13224 | 14566 | -0.107 | -0.0431 | No | ||
| 171 | PPP1R12C | 6115 17987 | 14668 | -0.112 | -0.0472 | No | ||
| 172 | TULP4 | 12740 | 14864 | -0.125 | -0.0562 | No | ||
| 173 | NSMAF | 15944 | 14894 | -0.127 | -0.0562 | No | ||
| 174 | PGAM1 | 9556 | 14932 | -0.130 | -0.0566 | No | ||
| 175 | SERTAD3 | 18327 | 15061 | -0.142 | -0.0618 | No | ||
| 176 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 15065 | -0.142 | -0.0602 | No | ||
| 177 | FASLG | 13789 | 15127 | -0.147 | -0.0616 | No | ||
| 178 | PIP5K3 | 14234 | 15311 | -0.165 | -0.0695 | No | ||
| 179 | IQWD1 | 13778 | 15608 | -0.205 | -0.0830 | No | ||
| 180 | ARRDC3 | 4191 | 15616 | -0.205 | -0.0808 | No | ||
| 181 | PIK3R3 | 5248 | 15646 | -0.210 | -0.0798 | No | ||
| 182 | INSIG1 | 6073 10525 10526 | 15665 | -0.212 | -0.0781 | No | ||
| 183 | HCN2 | 19965 | 15740 | -0.224 | -0.0793 | No | ||
| 184 | SGPP1 | 21039 | 15800 | -0.234 | -0.0796 | No | ||
| 185 | ANP32E | 15500 | 15898 | -0.250 | -0.0817 | No | ||
| 186 | APRIN | 4145 | 16131 | -0.301 | -0.0906 | No | ||
| 187 | GRIA3 | 24349 11996 2649 | 16136 | -0.302 | -0.0870 | No | ||
| 188 | PCAF | 5226 9532 5225 | 16150 | -0.306 | -0.0839 | No | ||
| 189 | TSC1 | 7235 | 16205 | -0.322 | -0.0828 | No | ||
| 190 | MEF2D | 9379 | 16214 | -0.323 | -0.0792 | No | ||
| 191 | EOMES | 4669 | 16242 | -0.329 | -0.0765 | No | ||
| 192 | CD69 | 16979 8717 | 16322 | -0.351 | -0.0764 | No | ||
| 193 | DNAJB12 | 7146 | 16422 | -0.379 | -0.0770 | No | ||
| 194 | RNF4 | 5385 9729 | 16610 | -0.431 | -0.0818 | No | ||
| 195 | NRF1 | 17499 | 16635 | -0.441 | -0.0776 | No | ||
| 196 | DYNLT3 | 24187 | 16800 | -0.512 | -0.0801 | No | ||
| 197 | GATA2 | 17370 | 16835 | -0.530 | -0.0753 | No | ||
| 198 | SLC9A1 | 16053 | 16838 | -0.531 | -0.0687 | No | ||
| 199 | SESN3 | 19561 | 17492 | -0.860 | -0.0934 | No | ||
| 200 | PAFAH1B1 | 1340 5220 9524 | 17724 | -1.001 | -0.0935 | No | ||
| 201 | POLK | 21384 | 17878 | -1.101 | -0.0880 | No | ||
| 202 | NELF | 12174 | 17917 | -1.138 | -0.0758 | No | ||
| 203 | DOCK9 | 8369 | 17999 | -1.208 | -0.0651 | No | ||
| 204 | CCNE2 | 4491 | 18137 | -1.408 | -0.0549 | No | ||
| 205 | NFAT5 | 3921 7037 12036 | 18335 | -1.759 | -0.0435 | No | ||
| 206 | BCL2L11 | 2790 14861 | 18415 | -2.056 | -0.0221 | No | ||
| 207 | NLK | 5179 5178 | 18533 | -2.631 | 0.0045 | No |